Directed evolution of the site specificity of Cre recombinase
- PMID: 11904359
- PMCID: PMC123623
- DOI: 10.1073/pnas.022039799
Directed evolution of the site specificity of Cre recombinase
Abstract
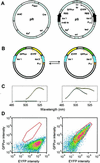
Cre recombinase from bacteriophage P1 recognizes a 34-bp recombination site, loxP, with exquisite sequence specificity and catalyzes the site-specific insertion, excision, or rearrangement of DNA. To better understand the molecular basis of protein-DNA recognition and generate recombinases with altered specificities, we have developed a directed evolution strategy that can be used to identify recombinases that recognize variant loxP sites. To be selected, members of a library of Cre variants produced by targeted random mutagenesis must rapidly catalyze recombination, in vivo, between two variant loxP sites that are located on a reporter plasmid. Recombination results in an altered pattern of fluorescent protein expression that can be identified by flow cytometry. Fluorescence-activated cell sorting can be used either to screen positively for recombinase variants that recognize a novel loxP site, or negatively for variants that cannot recognize the wild-type loxP site. The use of positive screening alone resulted in a relaxation of recombination site specificity, whereas a combination of positive and negative screening resulted in a switching of specificity. One of the identified recombinases selectively recombines a novel recombination site and operates at a rate identical to that of wild-type Cre. Analysis of the sequences of the resulting Cre variants provides insight into the evolution of these altered specificities. This and other systems should contribute to our understanding of protein-DNA recognition and may eventually be used to evolve custom-tailored recombinases that can be used for gene study and inactivation.
Figures

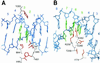



References
-
- Abremski K, Hoess R H, Sternberg N. Cell. 1983;32:1301–1311. - PubMed
-
- Abremski K, Hoess R. J Biol Chem. 1984;259:1509–1514. - PubMed
-
- Sternberg N, Hamilton D, Austin S, Yarmolinsky M, Hoess R. Cold Spring Harbor Symp Quant Biol. 1981;1:297–309. - PubMed
-
- Guo F, Gopaul D N, Van Duyne G D. Nature (London) 1997;389:40–46. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

