Fast 100-nm resolution three-dimensional microscope reveals structural plasticity of mitochondria in live yeast
- PMID: 11904401
- PMCID: PMC122530
- DOI: 10.1073/pnas.052545099
Fast 100-nm resolution three-dimensional microscope reveals structural plasticity of mitochondria in live yeast
Abstract
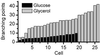
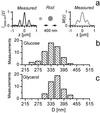
By introducing beam-scanning multifocal multiphoton 4Pi-confocal microscopy, we have attained fast fluorescence imaging of live cells with axial super resolution. Rapid scanning of up to 64 pairs of interfering high-angle fields and subsequent confocal detection enabled us to perform three to five times finer optical sectioning than confocal microscopy. In conjunction with nonlinear image restoration, we demonstrate, to our knowledge for the first time, three-dimensional imaging of live eukaryotic cells at an equilateral resolution of approximately 100 nm. This imaging mode allowed us to reveal the morphology and size of the green fluorescent protein-labeled mitochondrial compartment of live Saccharomyces cerevisiae (bakers' yeast) growing on different carbon sources. Our studies show that mitochondria of cells grown on medium containing glycerol as the only carbon source, as opposed to glucose-grown cells, exhibit a strongly branched tubular reticulum. We determine the average tubular diameter and find that it increases from 339 +/- 5 nm to 360 +/- 4 nm when changing from glucose to glycerol, that is, from a fermentable to a nonfermentable carbon source. Moreover, this change is associated with a 2.8-fold increase of the surface of the reticulum, resulting in an average increase in volume of the mitochondrial compartment by a factor of 3.0 +/- 0.2.
Figures





References
-
- Pawley J. Handbook of Biological Confocal Microscopy. New York: Plenum; 1995.
-
- Goodman J W. Introduction to Fourier Optics. New York: Mc Graw–Hill; 1968.
-
- Hell S W, Stelzer E H K. Opt Commun. 1992;93:277–282.
-
- Hell S W, Wichmann J. Opt Lett. 1994;19:780–782. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

