Elongator is a histone H3 and H4 acetyltransferase important for normal histone acetylation levels in vivo
- PMID: 11904415
- PMCID: PMC122555
- DOI: 10.1073/pnas.022042899
Elongator is a histone H3 and H4 acetyltransferase important for normal histone acetylation levels in vivo
Abstract
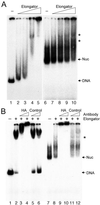
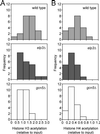
The elongating, hyperphosphorylated form of RNA polymerase II is associated with the Elongator complex, which has the histone acetyltransferase (HAT) Elp3 as a subunit. Here we show that, in contrast to the isolated Elp3 subunit, the activity of intact Elongator complex is directed specifically toward the amino-terminal tails of histone H3 and H4, and that Elongator can acetylate both core histones and nucleosomal substrates. The predominant acetylation sites are lysine-14 of histone H3 and lysine-8 of histone H4. The three smallest Elongator subunits--Elp4, Elp5, and Elp6--are required for HAT activity, and Elongator binds to both naked and nucleosomal DNA. By using chromatin immunoprecipitation, we show that the levels of multiply acetylated histone H3 and H4 in chromatin are decreased in vivo in yeast cells lacking ELP3.
Figures



References
-
- Kornberg R D, Lorch Y. Cell. 1999;98:285–294. - PubMed
-
- Kornberg R D, Lorch Y. Curr Opin Genet Dev. 1999;9:148–151. - PubMed
-
- Workman J L, Kingston R E. Annu Rev Biochem. 1998;67:545–579. - PubMed
-
- Stallcup M R. Oncogene. 2001;20:3014–3020. - PubMed
-
- Brownell J E, Zhou J, Ranalli T, Kobayashi R, Edmondson D G, Roth S Y, Allis C D. Cell. 1996;84:843–851. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

