Metabolic efficiency and amino acid composition in the proteomes of Escherichia coli and Bacillus subtilis
- PMID: 11904428
- PMCID: PMC122586
- DOI: 10.1073/pnas.062526999
Metabolic efficiency and amino acid composition in the proteomes of Escherichia coli and Bacillus subtilis
Abstract
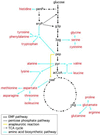
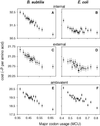
Biosynthesis of an Escherichia coli cell, with organic compounds as sources of energy and carbon, requires approximately 20 to 60 billion high-energy phosphate bonds [Stouthamer, A. H. (1973) Antonie van Leeuwenhoek 39, 545-565]. A substantial fraction of this energy budget is devoted to biosynthesis of amino acids, the building blocks of proteins. The fueling reactions of central metabolism provide precursor metabolites for synthesis of the 20 amino acids incorporated into proteins. Thus, synthesis of an amino acid entails a dual cost: energy is lost by diverting chemical intermediates from fueling reactions and additional energy is required to convert precursor metabolites to amino acids. Among amino acids, costs of synthesis vary from 12 to 74 high-energy phosphate bonds per molecule. The energetic advantage to encoding a less costly amino acid in a highly expressed gene can be greater than 0.025% of the total energy budget. Here, we provide evidence that amino acid composition in the proteomes of E. coli and Bacillus subtilis reflects the action of natural selection to enhance metabolic efficiency. We employ synonymous codon usage bias as a measure of translation rates and show increases in the abundance of less energetically costly amino acids in highly expressed proteins.
Figures


References
-
- Richmond R C. Nature (London) 1970;225:1025–1028. - PubMed
-
- King J L, Jukes T H. Science. 1969;164:788–797. - PubMed
-
- Doolittle R F. In: The Proteins. 3rd Ed. Neurath H, Hill R L, editors. Vol. 4. New York: Academic; 1979. pp. 1–118.
-
- Li W-H. Molecular Evolution. Sunderland, MA: Sinauer; 1997. pp. 185–190.
-
- Sueoka N. Cold Spring Harbor Symp Quant Biol. 1961;26:35–43. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

