Human immunodeficiency virus cDNA metabolism: notable stability of two-long terminal repeat circles
- PMID: 11907213
- PMCID: PMC136088
- DOI: 10.1128/jvi.76.8.3739-3747.2002
Human immunodeficiency virus cDNA metabolism: notable stability of two-long terminal repeat circles
Abstract
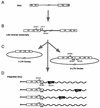
Early steps of retroviral replication involve reverse transcription of the viral RNA to yield a linear double-stranded cDNA copy and then integration of the viral cDNA into a chromosome of the host cell. A portion of the viral cDNA can also follow nonproductive pathways in which it becomes circularized. In one pathway, the ends of the linear cDNA become joined together by the cellular nonhomologous DNA end-joining system to form two-long terminal repeat (2-LTR) circles. It has been argued that 2-LTR circles are quickly degraded in human immunodeficiency virus (HIV)-infected cells, allowing the presence of 2-LTR circles to be used as a marker for ongoing de novo infection in patients. Following this idea, detection of 2-LTR circles in patients undergoing successful highly active antiretroviral therapy has led to the proposal that viral replication persists despite treatment. We have used fluorescence-monitored PCR (Taqman) to quantitate the metabolism of HIV cDNA early after infection. Contrary to previous work, we find that 2-LTR circles are actually quite stable in experiments where confounding variables are controlled. Thus, studies relying on the lability of 2-LTR circles are open to reinterpretation. We also used the quantitative PCR methods to analyze the effects of MG132, a proteasome inhibitor, which revealed that viral complexes containing mostly completed cDNAs are the primary substrates for proteasome-mediated degradation.
Figures




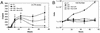
References
-
- Butler, S., M. Hansen, and F. D. Bushman. 2001. A quantitative assay for HIV cDNA integration in vivo. Nat. Med. 7:631-634. - PubMed
-
- Coffin, J. M., S. H. Hughes, and H. E. Varmus. 1997. Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. - PubMed
-
- Follenzi, A., L. E. Ailes, S. Bakovic, M. Gueuna, and L. Naldini. 2000. Gene transfer by lentiviral vectors is limited by nuclear translocation and rescued by HIV-1 pol sequences. Nat. Genet. 25:217-222. - PubMed
-
- Hansen, M. S. T., G. J. I. Smith, T. Kafri, V. Molteni, J. S. Siegel, and F. D. Bushman. 1999. Integration complexes derived from HIV vectors for rapid assays in vitro. Nat. Biotechnol. 17:578-582. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

