Persistent and transient replication of full-length hepatitis C virus genomes in cell culture
- PMID: 11907240
- PMCID: PMC136109
- DOI: 10.1128/jvi.76.8.4008-4021.2002
Persistent and transient replication of full-length hepatitis C virus genomes in cell culture
Abstract
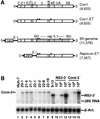
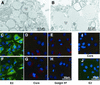
The recently developed subgenomic hepatitis C virus (HCV) replicons were limited by the fact that the sequence encoding the structural proteins was missing. Therefore, important information about a possible influence of these proteins on replication and pathogenesis and about the mechanism of virus formation could not be obtained. Taking advantage of three cell culture-adaptive mutations that enhance RNA replication synergistically, we generated selectable full-length HCV genomes that amplify to high levels in the human hepatoma cell line Huh-7 and can be stably propagated for more than 6 months. The structural proteins are efficiently expressed, with the viral glycoproteins E1 and E2 forming heterodimers which are stable under nondenaturing conditions. No disulfide-linked glycoprotein aggregates were observed, suggesting that the envelope proteins fold productively. Electron microscopy studies indicate that cell lines harboring these full-length HCV RNAs contain lipid droplets. The majority of the core protein was found on the surfaces of these structures, whereas the glycoproteins appear to localize to the endoplasmic reticulum and cis-Golgi compartments. In agreement with this distribution, no endoglycosidase H-resistant forms of these proteins were detectable. In a search for the production of viral particles, we noticed that these cells release substantial amounts of nuclease-resistant HCV RNA-containing structures with a buoyant density of 1.04 to 1.1 g/ml in iodixanol gradients. The same observation was made in transient-replication assays using an authentic highly adapted full-length HCV genome that lacks heterologous sequences. However, the fact that comparable amounts of such RNA-containing structures were found in the supernatant of cells carrying subgenomic replicons demonstrates a nonspecific release independent of the presence of the structural proteins. These results suggest that Huh-7 cells lack host cell factors that are important for virus particle assembly and/or release.
Figures




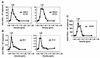
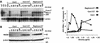
References
-
- Barba, G., F. Harper, T. Harada, M. Kohara, S. Goulinet, Y. Matsuura, G. Eder, Z. Schaff, M. J. Chapman, T. Miyamura, and C. Brechot. 1997. Hepatitis C virus core protein shows a cytoplasmic localization and associates to cellular lipid storage droplets. Proc. Natl. Acad. Sci. USA 94:1200-1205. - PMC - PubMed
-
- Barbaro, G., G. Di Lorenzo, A. Asti, M. Ribersani, G. Belloni, B. Grisorio, G. Filice, and G. Barbarini. 1999. Hepatocellular mitochondrial alterations in patients with chronic hepatitis C: ultrastructural and biochemical findings. Am. J. Gastroenterol. 94:2198-2205. - PubMed
-
- Bartenschlager, R. 1999. The NS3/4A proteinase of the hepatitis C virus: unravelling structure and function of an unusual enzyme and a prime target for antiviral therapy. J. Viral Hepatitis 10:1-2. - PubMed
-
- Bartenschlager, R., and V. Lohmann. 2000. Replication of hepatitis C virus. J. Gen. Virol. 81:1631-1648. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

