A natural intergenotypic recombinant of hepatitis C virus identified in St. Petersburg
- PMID: 11907242
- PMCID: PMC136067
- DOI: 10.1128/jvi.76.8.4034-4043.2002
A natural intergenotypic recombinant of hepatitis C virus identified in St. Petersburg
Abstract
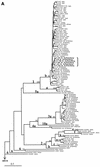
Hepatitis C virus (HCV) evolution is thought to proceed by mutations within the six genotypes. Here, we report on a viable spontaneous HCV recombinant and we show that recombination may play a role in the evolution of this virus. Previously, 149 HCV strains from St. Petersburg had been subtyped by limited sequencing within the NS5B region. In the present study, the core regions of 41 of these strains were sequenced to investigate the concordance of HCV genotyping for these two genomic regions. Two phylogenetically related HCV strains were found to belong to different subtypes, 2k and 1b, according to sequence analysis of the 5' untranslated region (5'UTR)-core and the NS5B regions, respectively. By sequencing of the E2-p7-NS2 region, the crossover point was mapped within the NS2 region, probably between positions 3175 and 3176 (according to the numbering system for strain pj6CF). Sequencing of the 5'UTR-core regions of four other HCV strains, phylogenetically related to the above-mentioned two strains (based on analysis within the NS5B region), revealed that these four strains were also recombinants. Since a nonrecombinant 2k strain was found in St. Petersburg, the recombination may have taken place there around a decade ago. Since the frequency of this recombinant is now high enough to allow the detection of the recombinant in a fraction of the city's population, it seems to be actively spreading there. The reported recombinant is tentatively designated RF1-2k/1b, in agreement with the nomenclature used for HIV recombinants. Recombination between HCV genotypes must now be considered in the classification, laboratory diagnosis, and treatment of HCV infection.
Figures





References
-
- De Francesco, F. R. 1999. Molecular virology of the hepatitis C virus. J. Hepatol. 31(Suppl. 1):47-53. - PubMed
-
- Dixit, V., S. Quan, P. Martin, D. Larson, M. Brezina, R. DiNello, K. Sra, J. Y. N. Lau, D. Chien, J. Kolberg, A. Tagger, G. Davis, A. Polito, and G. Gitnick. 1995. Evaluation of a novel serotyping system for hepatitis C virus: strong correlation with standard genotyping methodologies. J. Clin. Microbiol. 33:2978-2983. - PMC - PubMed
-
- Furione, M., S. Guillot, D. Otelea, J. Balanant, A. Candrea, and R. Crainic. 1993. Polioviruses with natural recombinant genomes isolated from vaccine-associated paralytic poliomyelitis. Virology 196:199-208. - PubMed
-
- Gale, M. J., M. J. Korth, N. M. Tang, S. L. Tan, D. A. Hopkins, T. E. Dever, S. J. Polyak, D. R. Gretch, and M. G. Katze. 1997. Evidence that hepatitis C virus resistance to interferon is mediated through repression of the PKR protein kinase by the nonstructural 5A protein. Virology 230:217-227. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

