Structural requirements for the assembly of Norwalk virus-like particles
- PMID: 11907243
- PMCID: PMC136079
- DOI: 10.1128/jvi.76.8.4044-4055.2002
Structural requirements for the assembly of Norwalk virus-like particles
Abstract
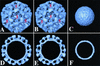
Norwalk virus (NV) is the prototype strain of a group of human caliciviruses responsible for epidemic outbreaks of acute gastroenteritis. While these viruses do not grow in tissue culture cells or animal models, expression of the capsid protein in insect cells results in the self-assembly of recombinant NV virus-like particles (rNV VLPs) that are morphologically and antigenically similar to native NV. The X-ray structure of the rNV VLPs has revealed that the capsid protein folds into two principal domains: a shell (S) domain and a protruding (P) domain (B. V. V. Prasad, M. E. Hardy, T. Dokland, J. Bella, M. G. Rossmann, and M. K. Estes, Science 286:287-290, 1999). To investigate the structural requirements for the assembly of rNV VLPs, we performed mutational analyses of the capsid protein. We examined the ability of 10 deletion mutants of the capsid protein to assemble into VLPs in insect cell cultures. Deletion of the N-terminal 20 residues, suggested by the X-ray structure to be involved in a switching mechanism during assembly, did not affect the ability of the mutant capsid protein to self-assemble into 38-nm VLPs with a T=3 icosahedral symmetry. Further deletions in the N-terminal region affected particle assembly. Deletions in the C-terminal regions of the P domain, involved in the interactions between the P and S domains, did not block the assembly process, but they affected the size and stability of the particles. Mutants carrying three internal deletion mutations in the P domain, involved in maintaining dimeric interactions, produced significantly larger 45-nm particles, albeit in low yields. The complete removal of the protruding domain resulted in the formation of smooth particles with a diameter that is slightly smaller than the 30-nm diameter expected from the rNV structure. These studies indicate that the shell domain of the NV capsid protein contains everything required to initiate the assembly of the capsid, whereas the entire protruding domain contributes to the increased stability of the capsid by adding intermolecular contacts between the dimeric subunits and may control the size of the capsid.
Figures




References
-
- Crowther, R. A. 1971. Procedures for three-dimensional reconstruction of spherical viruses by Fourier synthesis from electron micrographs. Phil. Trans. R. Scot. Lond. B Biol. Sci. 261:221-230. - PubMed
-
- Crowther, R. A., D. J. DeRosier, and A. Klug. 1970. The reconstruction of a three-dimensional structure from projections and its application to electron microscopy. Proc. R. Soc. Lond. 317:319. - PubMed
-
- Delgadillo, R. A., R. M. Smets, Q. R. Yang, D. Vanden Berghe, and A. Neetens. 1988. The use of iodinated density gradient medium for the isolation of rod outer segments. Experientia 44:702-704. - PubMed
-
- Dubochet, J., M. Adrian, J. J. Chang, J. C. Homo, J. Lepault, A. W. McDowall, and P. Schultz. 1988. Cryo-electron microscopy of vitrified specimens. Q. Rev. Biophys. 21:129-228. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

