Expression profile matrix of Arabidopsis transcription factor genes suggests their putative functions in response to environmental stresses
- PMID: 11910004
- PMCID: PMC150579
- DOI: 10.1105/tpc.010410
Expression profile matrix of Arabidopsis transcription factor genes suggests their putative functions in response to environmental stresses
Abstract
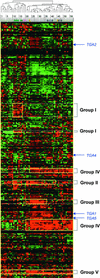
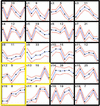
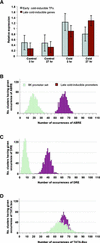
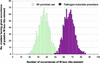
Numerous studies have shown that transcription factors are important in regulating plant responses to environmental stress. However, specific functions for most of the genes encoding transcription factors are unclear. In this study, we used mRNA profiles generated from microarray experiments to deduce the functions of genes encoding known and putative Arabidopsis transcription factors. The mRNA levels of 402 distinct transcription factor genes were examined at different developmental stages and under various stress conditions. Transcription factors potentially controlling downstream gene expression in stress signal transduction pathways were identified by observed activation and repression of the genes after certain stress treatments. The mRNA levels of a number of previously characterized transcription factor genes were changed significantly in connection with other regulatory pathways, suggesting their multifunctional nature. The expression of 74 transcription factor genes responsive to bacterial pathogen infection was reduced or abolished in mutants that have defects in salicylic acid, jasmonic acid, or ethylene signaling. This observation indicates that the regulation of these genes is mediated at least partly by these plant hormones and suggests that the transcription factor genes are involved in the regulation of additional downstream responses mediated by these hormones. Among the 43 transcription factor genes that are induced during senescence, 28 of them also are induced by stress treatment, suggesting extensive overlap responses to these stresses. Statistical analysis of the promoter regions of the genes responsive to cold stress indicated unambiguous enrichment of known conserved transcription factor binding sites for the responses. A highly conserved novel promoter motif was identified in genes responding to a broad set of pathogen infection treatments. This observation strongly suggests that the corresponding transcription factors play general and crucial roles in the coordinated regulation of these specific regulons. Although further validation is needed, these correlative results provide a vast amount of information that can guide hypothesis-driven research to elucidate the molecular mechanisms involved in transcriptional regulation and signaling networks in plants.
Figures


References
-
- Alonso, J.M., Hirayama, T., Roman, G., Nourizadeh, S., and Ecker, J.R. (1999). EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science 284, 2148–2152. - PubMed
-
- Borden, K.L. (2000). RING domains: Master builders of molecular scaffolds? J. Mol. Biol. 295, 1103–1112. - PubMed
-
- Cao, H., Glazebrook, J., Clarke, J.D., Volko, S., and Dong, X. (1997). The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88, 57–63. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

