The Cf-9 disease resistance protein is present in an approximately 420-kilodalton heteromultimeric membrane-associated complex at one molecule per complex
- PMID: 11910014
- PMCID: PMC150589
- DOI: 10.1105/tpc.010357
The Cf-9 disease resistance protein is present in an approximately 420-kilodalton heteromultimeric membrane-associated complex at one molecule per complex
Retraction in
-
RETRACTED: The Cf-9 Disease Resistance Protein Is Present in an ∼420-Kilodalton Heteromultimeric Membrane-Associated Complex at One Molecule per Complex.Plant Cell. 2017 Feb 23;29(3):600. doi: 10.1105/tpc.17.00150. Online ahead of print. Plant Cell. 2017. PMID: 28232383 Free PMC article. No abstract available.
Abstract
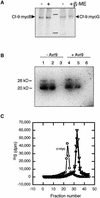
The tomato Cf-9 gene confers race-specific resistance to the fungal pathogen Cladosporium fulvum expressing the corresponding avirulence gene Avr9. In tobacco, Cf-9 confers a hypersensitive response to the Avr9 peptide. To investigate Cf-9 protein function in initiating defense signaling, we engineered a functional C-terminal fusion of the Cf-9 gene with the TAP (Tandem Affinity Purification) tag. In addition, we established a transient expression assay in Nicotiana benthamiana leaves for the production of functional Cf-9:myc and Cf-9:TAP. Transiently expressed Cf-9:myc and Cf-9:TAP proteins induced an Avr9-dependent hypersensitive response, consistent with previous results with stably transformed tobacco plants and derived cell suspension cultures expressing c-myc-tagged Cf-9. Gel filtration of microsomal fractions solubilized with octylglucoside revealed that the Cf-9 protein, either as c-myc or TAP fusions, migrated at a molecular mass of 350 to 475 kD. By using blue native gel electrophoresis, the molecular size was confirmed to be approximately 420 kD. Our results suggest that only one Cf-9 protein molecule is present in the Cf-9 complex and that Cf-9 is part of a membrane complex consisting of an additional glycoprotein partner(s). The high structural similarity between Cf proteins and Clavata2 (CLV2) of Arabidopsis, together with the similarity of molecular mass between Cf-9 and CLV complexes (420 and 450 kD, respectively), led us to investigate whether Cf-9 is integrated into membrane-associated protein complexes like those formed by CLV1 and CLV2. Unlike CLV2, the Cf-9 protein did not form disulfide-linked heterodimers, no ligand (Avr9)-dependent shift in the molecular mass of the Cf-9 complex was detected, and no Rho-GTPase-related proteins were found associated with Cf-9 under the conditions tested. Thus, Cf-9-dependent defense signaling and CLV2-dependent regulation of meristem development seem to be accomplished via distinct mechanisms, despite the structural similarity of their key components Cf-9 and CLV2.
Figures






References
-
- Arnold, I., Pfeiffer, K., Neupert, W., Stuart, R.A., and Schägger, H. (1999). ATP synthase of yeast mitochondria. J. Biol. Chem. 274, 36–40. - PubMed
-
- Blatt, M.R., Grabov, A., Brearley, J., Hammond-Kosack, K., and Jones, J.D.G. (1999). K+ channels of Cf-9-transgenic tobacco guard cells as targets for Cladosporium fulvum Avr9. Plant J. 19, 453–462. - PubMed
-
- Clark, S.E., Williams, R.W., and Meyerowitz, E.M. (1997). The Clavata1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell 89, 575–585. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

