Natively unfolded proteins: a point where biology waits for physics
- PMID: 11910019
- PMCID: PMC2373528
- DOI: 10.1110/ps.4210102
Natively unfolded proteins: a point where biology waits for physics
Abstract
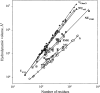
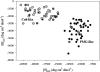
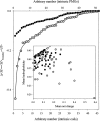
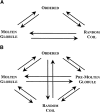
The experimental material accumulated in the literature on the conformational behavior of intrinsically unstructured (natively unfolded) proteins was analyzed. Results of this analysis showed that these proteins do not possess uniform structural properties, as expected for members of a single thermodynamic entity. Rather, these proteins may be divided into two structurally different groups: intrinsic coils, and premolten globules. Proteins from the first group have hydrodynamic dimensions typical of random coils in poor solvent and do not possess any (or almost any) ordered secondary structure. Proteins from the second group are essentially more compact, exhibiting some amount of residual secondary structure, although they are still less dense than native or molten globule proteins. An important feature of the intrinsically unstructured proteins is that they undergo disorder-order transition during or prior to their biological function. In this respect, the Protein Quartet model, with function arising from four specific conformations (ordered forms, molten globules, premolten globules, and random coils) and transitions between any two of the states, is discussed.
Figures


References
-
- Abercrombie, B.D., Kneale, G.G., Crane Robinson, C., Bradbury, E.M., Goodwin, G.H., Walker, J.M., and Johns, E.W. 1978. Studies on the conformational properties of the high-mobility-group chromosomal protein HMG 17 and its interaction with DNA. Eur. J. Biochem. 84 173–177. - PubMed
-
- Adler, A.J., Greenfield, N.J., and Fasman, G.D. 1973. Circular dichroism and optical rotatory dispersion of proteins and polypeptides. Methods Enzymol. 27 675–735. - PubMed
-
- Agianian, B., Leonard, K., Bonte, E., Van der Zandt, H., Becker, P.B., and Tucker, P.A. 1999. The glutamine-rich domain of the Drosophila GAGA factor is necessary for amyloid fiber formation in vitro, but not for chromatin remodelling. J. Mol. Biol. 285 527–544. - PubMed
-
- Alber, T., Gilbert, W.A., Ponzi, D.R., and Petsko, G.A. 1982. The role of mobility in the substrate binding and catalytic machinery of enzymes. Ciba Found. Symp. 93 4–24. - PubMed
-
- Alexandrescu, A.T., Abeygunawardana, C., and Shortle, D. 1994. Structure and dynamics of a denatured 131-residue fragment of staphylococcal nuclease: A heteronuclear NMR study. Biochemistry 33 1063–1072. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

