Differential recognition of surface proteins in Streptococcus pyogenes by two sortase gene homologs
- PMID: 11914350
- PMCID: PMC134975
- DOI: 10.1128/JB.184.8.2181-2191.2002
Differential recognition of surface proteins in Streptococcus pyogenes by two sortase gene homologs
Abstract
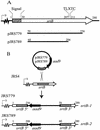
The interaction of Streptococcus pyogenes (group A streptococcus [GAS]) with its human host requires several surface proteins. In this study, we isolated mutations in a gene required for the surface localization of protein F by transposon mutagenesis of the M6 strain JRS4. This gene (srtA) encodes a protein homologous to Staphylococcus aureus sortase, which covalently links proteins containing an LPXTG motif to the cell wall. The GAS srtA mutant was defective in anchoring the LPXTG-containing proteins M6, protein F, ScpA, and GRAB to the cell surface. This phenotype was complemented when a wild-type srtA gene was provided in trans. The surface localization of T6, however, was unaffected by the srtA mutation. The M1 genome sequence contains a second open reading frame with a motif characteristic of sortase proteins. Inactivation of this gene (designated srtB) in strain JRS4 affected the surface localization of T6 but not M6, protein F, ScpA, or GRAB. This phenotype was complemented by srtB in trans. An srtA probe hybridized with DNA from all GAS strains tested (M types 1, 3, 4, 5, 6, 18, 22, and 50 and nontypeable strain 64/14) and from streptococcal groups C and G, while srtB hybridized with DNA from only a few GAS strains. We conclude that srtA and srtB encode sortase enzymes required for anchoring different subsets of proteins to the cell wall. It seems likely that the multiple sortase homologs in the genomes of other gram-positive bacteria have a similar substrate-specific role.
Figures






References
-
- Ashbaugh, C. D., T. J. Moser, M. H. Shearer, G. L. White, R. C. Kennedy, and M. R. Wessels. 2000. Bacterial determinants of persistent throat colonization and the associated immune response in a primate model of human group A streptococcal pharyngeal infection. Cell. Microbiol. 2:283-292. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

