Analysis of bacteria contaminating ultrapure water in industrial systems
- PMID: 11916667
- PMCID: PMC123900
- DOI: 10.1128/AEM.68.4.1548-1555.2002
Analysis of bacteria contaminating ultrapure water in industrial systems
Abstract
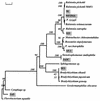
Bacterial populations inhabiting ultrapure water (UPW) systems were investigated. The analyzed UPW systems included pilot scale, bench scale, and full size UPW plants employed in the semiconductor and other industries. Bacteria present in the polishing loop of the UPW systems were enumerated by both plate counts and epifluorescence microscopy. Assessment of bacterial presence in UPW by epifluorescence microscopy (cyanotolyl tetrazolium chloride [CTC] and DAPI [4',6'-diamidino-2-phenylindole] staining) showed significantly higher numbers (10 to 100 times more bacterial cells were detected) than that determined by plate counts. A considerable proportion of the bacteria present in UPW (50 to 90%) were cells that did not give a positive signal with CTC stain. Bacteria isolated from the UPW systems were mostly gram negative, and several groups seem to be indigenous for all of the UPW production systems studied. These included Ralstonia pickettii, Bradyrhizobium sp., Pseudomonas saccharophilia, and Stenotrophomonas strains. These bacteria constituted a significant part of the total number of isolated strains (>or=20%). Two sets of primers specific to R. pickettii and Bradyrhizobium sp. were designed and successfully used for the detection of the corresponding bacteria in the concentrated UPW samples. Unexpectedly, nifH gene sequences were found in Bradyrhizobium sp. and some P. saccharophilia strains isolated from UPW. The widespread use of nitrogen gas in UPW plants may be associated with the presence of nitrogen-fixing genes in these bacteria.
Figures

References
-
- American Society for Testing and Materials. 2000. Standard test methods for microbiological monitoring of water used for processing electron and microelectronic devices by direct pressure tap sampling valve and by the presterilized plastic bag method (F-1094), p. 287-290. In Annual Book of ASTM Standards. American Society for Testing and Materials, West Conshohocken, Pa.
-
- American Society for Testing and Materials. 2000. Standard guide for ultrapure water used in the electronics and semiconductor industry (D-5127), p. 495-499. In Annual Book of ASTM Standards. American Society for Testing and Materials, West Conshohocken, Pa.
-
- Barraquio, W. L., B. C. Parde, Jr., I. Watanabe, and R. Knowles. 1986. Nitrogen fixation by Pseudomonas saccharophila Doudoroff ATCC 15946. J. Gen. Microbiol. 132:237-241.
-
- Chan, Y.-K., W. L. Barraquio, and R. Knowles. 1994. N2-fixing pseudomonads and related soil bacteria. FEMS Microbiol. Rev. 13:95-118.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

