Mammalian class E vps proteins recognize ubiquitin and act in the removal of endosomal protein-ubiquitin conjugates
- PMID: 11916981
- PMCID: PMC2173266
- DOI: 10.1083/jcb.200112080
Mammalian class E vps proteins recognize ubiquitin and act in the removal of endosomal protein-ubiquitin conjugates
Abstract
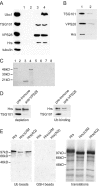
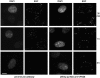
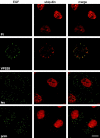
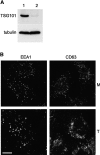
There is increasing evidence that ubiquitination of receptors provides an important endosomal sorting signal. Here we report that mammalian class E vacuolar protein-sorting (vps) proteins recognize ubiquitin. Both tumor susceptibility gene 101 (TSG101)/human VPS (hVPS)28 and hepatocyte growth factor receptor substrate (Hrs) cytosolic complexes bind ubiquitin-agarose. TSG101 and hVPS28 are localized to endosomes that contain internalized EGF receptor and label strongly for ubiquitinated proteins. Microinjection of anti-hVPS28 specifically retards EGF degradation and leads to endosomal accumulation of ubiquitin-protein conjugates. Likewise, depletion of TSG101 impairs EGF trafficking and causes dramatic relocalization of ubiquitin to endocytic compartments. Similar defects are found in cells overexpressing Hrs, further emphasizing the links between class E protein function, receptor trafficking, and endosomal ubiquitination.
Figures







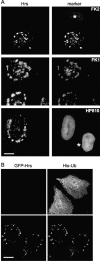
References
-
- Babst, M., G. Odorizzi, E.J. Estepa, and S.D. Emr. 2000. Mammalian tumor susceptibility gene 101 (TSG101) and the yeast homologue, Vps23p, both function in late endosomal trafficking. Traffic. 1:248–258. - PubMed
-
- Bishop, N., and P. Woodman. 2001. Tsg101/mammalian vps23 and mammalian vps28 interact directly and are recruited to vps4-induced endosomes. J. Biol. Chem. 276:11735–11742. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

