A cycle of Vam7p release from and PtdIns 3-P-dependent rebinding to the yeast vacuole is required for homotypic vacuole fusion
- PMID: 11916982
- PMCID: PMC2173272
- DOI: 10.1083/jcb.200112098
A cycle of Vam7p release from and PtdIns 3-P-dependent rebinding to the yeast vacuole is required for homotypic vacuole fusion
Abstract
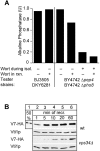
Vacuole fusion requires a coordinated cascade of priming, docking, and fusion. SNARE proteins have been implicated in the fusion itself, although their precise role in the cascade remains unclear. We now report that the vacuolar SNAP-23 homologue Vam7p is a mobile element of the SNARE complex, which moves from an initial association with the cis-SNARE complex via a soluble intermediate to the docking site. Soluble Vam7p is specifically recruited to vacuoles and can rescue a fusion reaction poisoned with antibodies to Vam7p. Both the recombinant Vam7p PX domain and a FYVE domain construct of human Hrs block the recruitment of Vam7p and vacuole fusion, demonstrating that phosphatidylinositol 3-phosphate is a primary receptor of Vam7p on vacuoles. We propose that the Vam7p cycle is linked to the availability of a lipid domain on yeast vacuoles, which is essential for coordinating the fusion reaction prior to and beyond docking.
Figures





References
-
- Burd, C.G., and S.D. Emr. 1998. Phosphatidylinositol(3)-phosphate signaling mediated by specific binding to RING FYVE domains. Mol. Cell. 2:157–162. - PubMed
-
- Cheever, M.L., T.K. Sato, T. de Beer, T.G. Kutateladze, S.D. Emr, and M. Overduin. 2001. Phox domain interaction with PtdIns(3)P targets the Vam7 t-SNARE to vacuole membranes. Nat. Cell Biol. 3:613–618. - PubMed
-
- Chen, Y.A., and R.H. Scheller. 2001. SNARE-mediated membrane fusion. Nat. Rev. Mol. Cell Biol. 2:98–106. - PubMed
-
- Chen, Y.A., S.J. Scales, S.M. Patel, Y.C. Doung, and R.H. Scheller. 1999. SNARE complex formation is triggered by Ca2+ and drives membrane fusion. Cell. 97:165–174. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

