SmpB functions in various steps of trans-translation
- PMID: 11917023
- PMCID: PMC101834
- DOI: 10.1093/nar/30.7.1620
SmpB functions in various steps of trans-translation
Abstract
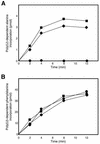
tmRNA has a dual function as a tRNA and an mRNA to facilitate trans-translation, in which a ribosome can switch between translation of a truncated mRNA and the tmRNA's tag sequence. SmpB is a tmRNA binding protein that has been identified to be essential for trans-translation in vivo. To further study the function of SmpB, an S30 fraction from an Escherichia coli strain, in which the set of genes for SmpB and tmRNA has been deleted from the genome, and His-tagged SmpB active in trans-translation were prepared. The SmpB-depleted S30 fraction had an ability to facilitate poly(U)-dependent tag-peptide synthesis in vitro when purified His-tagged SmpB was exogenously added together with tmRNA, although SmpB was not required for in vitro poly(U)-dependent poly(Phe) synthesis. It was also found that depletion of SmpB leads to a decrease in the level of tmRNA in the cell. In addition, SmpB considerably enhanced the aminoacylation of tmRNA by alanyl-tRNA synthetase in vitro. The aminoacylation enhancement by SmpB, the binding of SmpB to tmRNA and the effect of depletion of SmpB on the expression level of tmRNA in the cell were all affected by some mutations in the tRNA-like domain which cause a defect in ribosome binding leading to a trans-translation deficiency. These results demonstrate that, via binding to the tRNA-like domain of tmRNA, SmpB plays various roles: rescuing the tmRNA molecule from degradation in the cell, enhancing the aminoacylation of tmRNA and mediating the binding of tmRNA to ribosome.
Figures





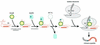
References
-
- Karzai A.W., Roche,E.D. and Sauer,R.T. (2000) The SsrA–SmpB system for protein tagging, directed degradation and ribosome rescue. NatureStruct. Biol., 7, 449–455. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

