Response of human REV1 to different DNA damage: preferential dCMP insertion opposite the lesion
- PMID: 11917024
- PMCID: PMC101843
- DOI: 10.1093/nar/30.7.1630
Response of human REV1 to different DNA damage: preferential dCMP insertion opposite the lesion
Abstract
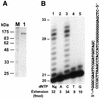
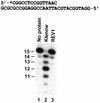
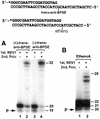
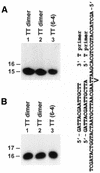
REV1 functions in the DNA polymerase zeta mutagenesis pathway. To help understand the role of REV1 in lesion bypass, we have examined activities of purified human REV1 opposite various template bases and several different DNA lesions. Lacking a 3'-->5' proofreading exonuclease activity, purified human REV1 exhibited a DNA polymerase activity on a repeating template G sequence, but catalyzed nucleotide insertion with 6-fold lower efficiency opposite a template A and 19-27-fold lower efficiency opposite a template T or C. Furthermore, dCMP insertion was greatly preferred regardless of the specific template base. Human REV1 inserted a dCMP efficiently opposite a template 8-oxoguanine, (+)-trans-anti-benzo[a]pyrene-N2-dG, (-)-trans-anti-benzo[a]pyrene-N2-dG and 1,N6-ethenoadenine adducts, very inefficiently opposite an acetylaminofluorene-adducted guanine, but was unresponsive to a template TT dimer or TT (6-4) photoproduct. Surprisingly, the REV1 specificity of nucleotide insertion was very similar in response to different DNA lesions with greatly preferred C insertion and least frequent A insertion. By combining the dCMP insertion activity of human REV1 with the extension synthesis activity of human polymerase kappa, bypass of the trans-anti-benzo[a]pyrene-N2-dG adducts and the 1,N6-ethenoadenine lesion was achieved by the two-polymerase two-step mechanism. These results suggest that human REV1 is a specialized DNA polymerase that may contribute to dCMP insertion opposite many types of DNA damage during lesion bypass.
Figures
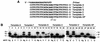
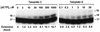

References
-
- Ohmori H., Friedberg,E.C., Fuchs,R.P.P., Goodman,M.F., Hanaoka,F., Hinkle,D., Kunkel,T.A., Lawrence,C.W., Livneh,Z., Nohmi,T., Prakash,L., Prakash,S., Todo,T., Walker,G.C., Wang,Z. and Woodgate,R. (2001) The Y-family of DNA polymerases. Mol. Cell, 8, 7–8. - PubMed
-
- Wang Z. (2001) Translesion synthesis by the UmuC family of DNA polymerases. Mutat. Res., 486, 59–70. - PubMed
-
- Nelson J.R., Lawrence,C.W. and Hinkle,D.C. (1996) Deoxycytidyl transferase activity of yeast REV1 protein. Nature, 382, 729–731. - PubMed
-
- Gibbs P.E. and Lawrence,C.W. (1995) Novel mutagenic properties of abasic sites in Saccharomyces cerevisiae. J. Mol. Biol., 251, 229–236. - PubMed
-
- Nelson J.R., Gibbs,P.E., Nowicka,A.M., Hinkle,D.C. and Lawrence,C.W. (2000) Evidence for a second function for Saccharomyces cerevisiae Rev1p. Mol. Microbiol., 37, 549–554. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

