The terminase subunits pUL56 and pUL89 of human cytomegalovirus are DNA-metabolizing proteins with toroidal structure
- PMID: 11917032
- PMCID: PMC101837
- DOI: 10.1093/nar/30.7.1695
The terminase subunits pUL56 and pUL89 of human cytomegalovirus are DNA-metabolizing proteins with toroidal structure
Abstract
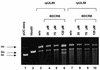
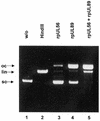
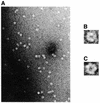
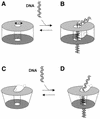
Herpesvirus DNA packaging involves binding and cleavage of DNA containing the specific DNA-packaging motifs. Here we report a first characterization of the terminase subunits pUL56 and pUL89 of human cytomegalovirus (HCMV). Both gene products were shown to have comparable nuclease activities in vitro. Under limiting protein concentrations the nuclease activity is enhanced by interaction of pUL56 and pUL89. High amounts of 2-bromo-5,6-dichloro-1-beta-D-ribofuranosyl benzimidazole partially inhibited the pUL89-associated nuclease activity. It was demonstrated that pUL56 is able to bind to nucleocapsids in vivo. Electron microscopy (EM) and image analysis of purified pUL56 revealed that the molecules occurred as a distinct ring-shaped structure with a pronounced cleft. EM analysis of purified pUL89 demonstrated that this protein is also a toroidal DNA-metabolizing protein. Upon interaction of pUL56 with linearized DNA, the DNA remains uncut while the cutting event itself is mediated by pUL89. Using biochemical assays in conjunction with EM pUL56 was shown to (i) bind to DNA and (ii) associate with the capsid. In contrast to this, EM analysis implied that pUL89 is required to effect DNA cleavage. The data provide the first insights into the terminase-dependent viral DNA-packaging mechanism of HCMV.
Figures







References
-
- Chou J. and Roizman,B. (1985) The isomerization of the herpes simplex virus 1 genome: identification of the cis-acting and recombination sites within the domain of the a sequence. Cell, 41, 803–811. - PubMed
-
- Black L.W. (1988) DNA packaging in dsDNA bacteriophages. In Calendar,R. (ed.), The Bacteriophages. Plenum, New York, NY, pp. 321–373.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

