Structural analysis of conserved base pairs in protein-DNA complexes
- PMID: 11917033
- PMCID: PMC101836
- DOI: 10.1093/nar/30.7.1704
Structural analysis of conserved base pairs in protein-DNA complexes
Abstract
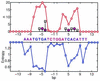
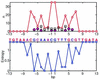
Understanding of protein-DNA interactions is crucial for prediction of DNA-binding specificity of transcription factors and design of novel DNA-binding proteins. In this paper we develop a novel approach to analysis of protein-DNA interactions. We bring together two sources of information: (i) structures of protein-DNA complexes (PDB/NDB database) and (ii) experimentally obtained sites recognized by DNA-binding proteins. Sites are used to compute conservation (information content) of each base pair, which indicates relative importance of the base pair in specific recognition. The main result of this study is that conservation of base pairs in a site exhibits significant correlation with the number of contacts the base pairs have with the protein. In particular, base pairs that have more contacts with the protein are more conserved in evolution. As natural as it is, this result has never been reported before. We also observe that for most of the studied proteins, hydrogen bonds and hydrophobic interactions alone cannot explain the pattern of evolutionary conservation in the binding site suggesting cumulative contribution of different types of interactions to specific recognition. Implications for prediction of the DNA-binding specificity are discussed.
Figures


References
-
- Stover C., Pham,X., Erwin,A., Mizoguchi,S., Warrener,P., Hickey,M., Brinkman,F., Hufnagle,W., Kowalik,D., Lagrou,M., Garber,R., Goltry,L., Tolentino,E., Westbrock-Wadman,S., Yuan,Y., Brody,L., Coulter,S., Folger,K., Kas,A., Larbig,K., Lim,R., Smith,K., Spencer,D., Wong,G., Wu,Z. and Paulsen,I. (2000) Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature, 406, 959–664. - PubMed
-
- Berman H., Zardecki,C. and Westbrook,J. (1998) The nucleic acid database: a resource for nucleic acid science. Acta Crystallogr. D. Biol. Crystallogr., 54, 1095–1104. - PubMed
-
- Larson C. and Verdine,G. (1996) The chemistry of protein-DNA interactions. In Hecht,S.M. (ed.), Bioorganic Chemistry: Nucleic Acids. Oxford University Press, Oxford, UK, pp. 324–346.
-
- Fields D., He,Y., Al-Uzri,A. and Stormo,G. (1997) Quantitative specificity of the Mnt repressor. J. Mol. Biol., 271, 178–194. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

