Mitochondrial DNA restriction fragment length polymorphism (RFLP) and 18S small-subunit ribosomal DNA PCR-RFLP analyses of Acanthamoeba isolated from contact lens storage cases of residents in southwestern Korea
- PMID: 11923331
- PMCID: PMC140361
- DOI: 10.1128/JCM.40.4.1199-1206.2002
Mitochondrial DNA restriction fragment length polymorphism (RFLP) and 18S small-subunit ribosomal DNA PCR-RFLP analyses of Acanthamoeba isolated from contact lens storage cases of residents in southwestern Korea
Abstract
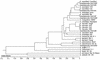
We applied ribosomal DNA PCR-restriction fragment length polymorphism (RFLP) and mitochondrial DNA (mtDNA) RFLP analyses to 43 Acanthamoeba environmental isolates (KA/LH1 to KA/LH43) from contact lens storage cases in southwestern Korea. These isolates were compared to American Type Culture Collection strains and clinical isolates (KA/E1 to KA/E12) from patients with keratitis. Seven riboprint patterns were seen. To identify the species of the isolates, a phylogenetic tree was constructed based on the comparison of riboprint patterns with reference strains. Four types accounted for 39 of the isolates belonging to the A. castellanii complex. The most predominant (48.8%) type was A. castellanii KA/LH2 type, which had identical riboprint and mtDNA RFLP patterns to those of A. castellanii Castellani, KA/E3 and KA/E8. The riboprint and mtDNA RFLP patterns of the KA/LH7 (20.9%) type were identical to those of A. castellanii Ma, a corneal isolate from the United States. The riboprint and mtDNA RFLP patterns of the KA/LH1 (18.6%) type were the same as those of A. lugdunensis L3a, KA/E2, and KA/E12. The prevalent pattern for each type of Acanthamoeba in southwestern Korea was very different from that from southeastern Korea and Seoul, Korea. It is noteworthy that 38 (88.4%) out of 43 isolates from contact lens storage cases of the residents in southwestern Korea revealed mtDNA RFLP and riboprint patterns identical to those found for clinical isolates in our area. This indicates that most isolates from contact lens storage cases in the surveyed area are potential keratopathogens. More attention should be paid to the disinfection of contact lens storage cases to prevent possible amoebic keratitis.
Figures



References
-
- Bhattacharya, D., T. Helmchen, and M. Melkonian. 1998. Molecular evolutionary analyses of nuclear-encoded small subunit ribosomal RNA identify and independent rhizopod lineage containing the Euglyphina and the Chlorarachniophyta. J. Eukaryot. Microbiol. 42:65-69. - PubMed
-
- Bogler, S. A., C. D. Zarley, L. L. Burianek, P. A. Fuerst, and T. J. Byers. 1983. Interstrain mitochondrial DNA polymorphism detected in Acanthamoeba by restriction endonuclease analysis. Mol. Biochem. Parasitol. 8:145-163. - PubMed
-
- Byers, T. J., S. A. Bogler, and L. L. Burianek. 1983. Analysis of mitochondrial DNA variation as an approach to systematic relationships in the genus Acanthamoeba. J. Protozool. 30:198-203.
-
- Byers, T. J., E. R. Hugo, and V. J. Stewart. 1990. Genes of Acanthamoeba: DNA, RNA and protein sequence. J. Protozool. 37:s17-s25. - PubMed
-
- Chung, D. I., H. S. Yu, M. Y. Hwang, T. H. Kim, T. O. Kim, H. C. Yun, and H. H. Kong. 1998. Subgenus classification of Acanthamoeba by riboprinting. Korean J. Parasitol. 36:69-80. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

