Impaired postnatal hepatocyte proliferation and liver regeneration in mice lacking c-jun in the liver
- PMID: 11927562
- PMCID: PMC125360
- DOI: 10.1093/emboj/21.7.1782
Impaired postnatal hepatocyte proliferation and liver regeneration in mice lacking c-jun in the liver
Abstract
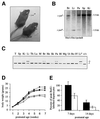
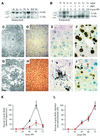
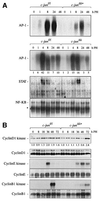
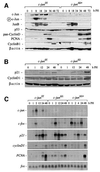
Mice lacking the AP-1 transcription factor c-jun die at mid-gestation showing heart defects and impaired hepatogenesis. To inactivate c-jun in hepatocytes, mice carrying a floxed c-jun allele were generated. Perinatal liver-specific c-jun deletion caused reduced hepatocyte proliferation and decreased body size. After partial hepatectomy, half of the mutants died and liver regeneration was impaired. This phenotype was not present in mice lacking the N-terminal phosphorylation sites of c-Jun. The failure to regenerate was accompanied by increased cell death and lipid accumulation in hepatocytes. Moreover, cyclin-dependent kinases and several cell cycle regulators were affected, resulting in inefficient G(1)-S phase progression. These studies identify c-Jun as a critical regulator of hepatocyte proliferation and survival during liver development and regeneration.
Figures


References
-
- Angel P., Hattori,K., Smeal,T. and Karin,M. (1988) The jun proto-oncogene is positively autoregulated by its product, Jun/AP1. Cell, 55, 875–885. - PubMed
-
- Arias J., Alberts,A.S., Brindle,P., Claret,F.X., Smeal,T., Karin,M., Feramisco,J. and Montminy,M. (1994) Activation of cAMP and mitogen responsive genes relies on a common nuclear factor. Nature, 370, 226–229. - PubMed
-
- Bannister A.J., Oehler,T., Wilhelm,D., Angel,P. and Kouzarides,T. (1995) Stimulation of c-Jun activity by CBP: c-Jun residues Ser63/73 are required for CBP induced stimulation in vivo and CBP binding in vitro. Oncogene, 11, 2509–2514. - PubMed
-
- Behrens A., Sibilia,M. and Wagner,E.F. (1999) Amino-terminal phosphorylation of c-Jun regulates stress-induced apoptosis and cellular proliferation. Nature Genet., 21, 326–329. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

