Genes directly controlled by CtrA, a master regulator of the Caulobacter cell cycle
- PMID: 11930012
- PMCID: PMC123699
- DOI: 10.1073/pnas.062065699
Genes directly controlled by CtrA, a master regulator of the Caulobacter cell cycle
Abstract
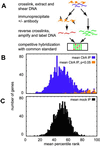
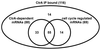
Studies of the genetic network that controls the Caulobacter cell cycle have identified a response regulator, CtrA, that controls, directly or indirectly, one-quarter of the 553 cell cycle-regulated genes. We have performed in vivo genomic binding site analysis of the CtrA protein to identify which of these genes have regulatory regions bound directly by CtrA. By combining these data with previous global analysis of cell cycle transcription patterns and gene expression profiles of mutant ctrA strains, we have determined that CtrA directly regulates at least 95 genes. The total group of CtrA-regulated genes includes those involved in polar morphogenesis, DNA replication initiation, DNA methylation, cell division, and cell wall metabolism. Also among the genes in this notably large regulon are 14 that encode regulatory proteins, including 10 two-component signal transduction regulatory proteins. Identification of additional regulatory genes activated by CtrA will serve to directly connect new regulatory modules to the network controlling cell cycle progression.
Figures



References
-
- Cho R J, Huang M, Campbell M J, Dong H, Steinmetz L, Sapinoso L, Hampton G, Elledge S J, Davis R W, Lockhart D J. Nat Genet. 2001;27:48–54. - PubMed
-
- Cho R J, Campbell M J, Winzeler E A, Steinmetz L, Conway A, Wodicka L, Wolfsberg T G, Gabrielian A E, Landsman D, Lockhart D J, Davis R W. Mol Cell. 1998;2:65–73. - PubMed
-
- Laub M T, McAdams H H, Feldblyum T, Fraser C M, Shapiro L. Science. 2000;290:2144–2148. - PubMed
-
- Quon K C, Marczynski G T, Shapiro L. Cell. 1996;84:83–93. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

