The complete genome of hyperthermophile Methanopyrus kandleri AV19 and monophyly of archaeal methanogens
- PMID: 11930014
- PMCID: PMC123701
- DOI: 10.1073/pnas.032671499
The complete genome of hyperthermophile Methanopyrus kandleri AV19 and monophyly of archaeal methanogens
Abstract
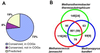
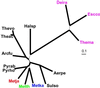
We have determined the complete 1,694,969-nt sequence of the GC-rich genome of Methanopyrus kandleri by using a whole direct genome sequencing approach. This approach is based on unlinking of genomic DNA with the ThermoFidelase version of M. kandleri topoisomerase V and cycle sequencing directed by 2'-modified oligonucleotides (Fimers). Sequencing redundancy (3.3x) was sufficient to assemble the genome with less than one error per 40 kb. Using a combination of sequence database searches and coding potential prediction, 1,692 protein-coding genes and 39 genes for structural RNAs were identified. M. kandleri proteins show an unusually high content of negatively charged amino acids, which might be an adaptation to the high intracellular salinity. Previous phylogenetic analysis of 16S RNA suggested that M. kandleri belonged to a very deep branch, close to the root of the archaeal tree. However, genome comparisons indicate that, in both trees constructed using concatenated alignments of ribosomal proteins and trees based on gene content, M. kandleri consistently groups with other archaeal methanogens. M. kandleri shares the set of genes implicated in methanogenesis and, in part, its operon organization with Methanococcus jannaschii and Methanothermobacter thermoautotrophicum. These findings indicate that archaeal methanogens are monophyletic. A distinctive feature of M. kandleri is the paucity of proteins involved in signaling and regulation of gene expression. Also, M. kandleri appears to have fewer genes acquired via lateral transfer than other archaea. These features might reflect the extreme habitat of this organism.
Figures

References
-
- Huber R, Kurr M, Jannasch H W, Stetter K O. Nature (London) 1989;342:833–836.
-
- Kurr M, Huber R, Konig H, Jannasch H W, Fricke H, Trincone A, Kristjansson J K, Stetter K O. Arch Microbiol. 1991;156:239–247.
-
- Hafenbradl D, Keller M, Thiericke R, Stetter K O. Syst Appl Microbiol. 1993;16:165–169.
-
- Shima S, Herault D A, Berkessel A, Thauer R K. Arch Microbiol. 1998;170:469–472. - PubMed
Publication types
MeSH terms
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

