Aminoadipate reductase gene: a new fungal-specific gene for comparative evolutionary analyses
- PMID: 11931673
- PMCID: PMC103663
- DOI: 10.1186/1471-2148-2-6
Aminoadipate reductase gene: a new fungal-specific gene for comparative evolutionary analyses
Abstract
Background: In fungi, aminoadipate reductase converts 2-aminoadipate to 2-aminoadipate 6-semialdehyde. However, other organisms have no homologue to the aminoadipate reductase gene and this pathway appears to be restricted to fungi. In this study, we designed degenerate primers for polymerase chain reaction (PCR) amplification of a large fragment of the aminoadipate reductase gene for divergent fungi.
Results: Using these primers, we amplified DNA fragments from the archiascomycetous yeast Saitoella complicata and the black-koji mold Aspergillus awamori. Based on an alignment of the deduced amino acid sequences, we constructed phylogenetic trees. These trees are consistent with current ascomycete systematics and demonstrate the potential utility of the aminoadipete reductase gene for phylogenetic analyses of fungi.
Conclusions: We believe that the comparison of aminoadipate reductase among species will be useful for molecular ecological and evolutionary studies of fungi, because this enzyme-encoding gene is a fungal-specific gene and generally appears to be single copy.
Figures
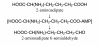

References
-
- Hawksworth DL, Rossman AY. Where are all the undescribed fungi? Phytopathology. 1997;87:888–891. - PubMed
-
- Nishida H, Sugiyama J. Phylogenetic relationships among Taphrina, Saitoella, and other higher fungi. Mol Biol Evol. 1993;10:431–436. - PubMed
-
- Nishida H, Sugiyama J. Archiascomycetes: detection of a major new lineage within the Ascomycota. Mycoscience. 1994;35:361–366.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Medical

