Avian bic, a gene isolated from a common retroviral site in avian leukosis virus-induced lymphomas that encodes a noncoding RNA, cooperates with c-myc in lymphomagenesis and erythroleukemogenesis
- PMID: 11932393
- PMCID: PMC155062
- DOI: 10.1128/jvi.76.9.4275-4286.2002
Avian bic, a gene isolated from a common retroviral site in avian leukosis virus-induced lymphomas that encodes a noncoding RNA, cooperates with c-myc in lymphomagenesis and erythroleukemogenesis
Abstract
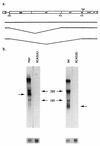
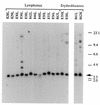
bic is a novel gene identified at a common retroviral integration site in avian leukosis virus-induced lymphomas and has been implicated as a collaborator with c-myc in B lymphomagenesis. It lacks an extensive open reading frame and is believed to function as an untranslated RNA (W. Tam, Gene 274:157-167, 2001; W. Tam, D. Ben-Yehuda, and W. S. Hayward, Mol. Cell. Biol. 17:1490-1502, 1997). The oncogenic potential of bic, particularly its ability to cooperate with c-myc in oncogenesis, was tested directly by expressing c-myc and bic, either singly or in pairwise combination, in cultured chicken embryo fibroblasts (CEFs) and in chickens using replication-competent retrovirus vectors. Coexpression of c-myc and bic in CEFs caused growth enhancement of cells. Most importantly, chick oncogenicity assays demonstrated that bic can cooperate with c-myc in lymphomagenesis and erythroleukemogenesis. The present study provides direct evidence for the involvement of untranslated RNAs in oncogenesis and provides further support for the role of noncoding RNAs as riboregulators.
Figures





References
-
- Adams, J. M., and S. Cory. 1992. Oncogene cooperation in leukaemogenesis. Cancer Surv. 15:119-141. - PubMed
-
- Adams, J. M., A. W. Harris, A. Strasser, S. Ogilvy, and S. Cory. 1999. Transgenic models of lymphoid neoplasia and development of a pan-hematopoietic vector. Oncogene 18:5268-5277. - PubMed
-
- Alexander, W. S., O. Bernard, S. Cory, and J. M. Adams. 1989. Lymphomagenesis in E mu-myc transgenic mice can involve ras mutations. Oncogene 4:575-581. - PubMed
-
- Askew, D. S., and F. Xu. 1999. New insights into the function of noncoding RNA and its potential role in disease pathogenesis. Histol. Histopathol. 14:235-241. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

