Sources of variability and effect of experimental approach on expression profiling data interpretation
- PMID: 11936955
- PMCID: PMC65691
- DOI: 10.1186/1471-2105-3-4
Sources of variability and effect of experimental approach on expression profiling data interpretation
Abstract
Background: We provide a systematic study of the sources of variability in expression profiling data using 56 RNAs isolated from human muscle biopsies (34 Affymetrix MuscleChip arrays), and 36 murine cell culture and tissue RNAs (42 Affymetrix U74Av2 arrays).
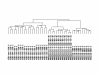
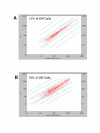
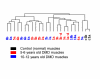
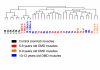
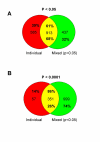
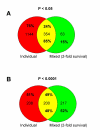
Results: We studied muscle biopsies from 28 human subjects as well as murine myogenic cell cultures, muscle, and spleens. Human MuscleChip arrays (4,601 probe sets) and murine U74Av2 Affymetrix microarrays were used for expression profiling. RNAs were profiled both singly, and as mixed groups. Variables studied included tissue heterogeneity, cRNA probe production, patient diagnosis, and GeneChip hybridizations. We found that the greatest source of variability was often different regions of the same patient muscle biopsy, reflecting variation in cell type content even in a relatively homogeneous tissue such as muscle. Inter-patient variation was also very high (SNP noise). Experimental variation (RNA, cDNA, cRNA, or GeneChip) was minor. Pre-profile mixing of patient cRNA samples effectively normalized both intra- and inter-patient sources of variation, while retaining a high degree of specificity of the individual profiles (86% of statistically significant differences detected by absolute analysis; and 85% by a 4-pairwise comparison survival method).
Conclusions: Using unsupervised cluster analysis and correlation coefficients of 92 RNA samples on 76 oligonucleotide microarrays, we found that experimental error was not a significant source of unwanted variability in expression profiling experiments. Major sources of variability were from use of small tissue biopsies, particularly in humans where there is substantial inter-patient variability (SNP noise).
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

