Generating segmental mutations in haloalkane dehalogenase: a novel part in the directed evolution toolbox
- PMID: 11937643
- PMCID: PMC113230
- DOI: 10.1093/nar/30.8.e35
Generating segmental mutations in haloalkane dehalogenase: a novel part in the directed evolution toolbox
Abstract
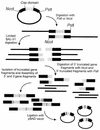
Directed evolution techniques allow us to genuinely mimic molecular evolution in vitro. To enhance this imitation of natural evolutionary processes on a laboratory scale in even more detail, we developed an in vitro method for the generation of random deletions and repeats. The pairwise fusion of two fragments of the same gene that are truncated by exonuclease BAL-31 either at the 3' or 5' side results in a deletion or a repeat at the fusion point. Although in principle the method randomly covers the whole gene, it can also be limited to a predefined area in the sequence by controlling the level of the initial truncation. To test the procedure and to illustrate its potential, we used haloalkane dehalogenase from Xanthobacter autotrophicus GJ10 (DhlA) as a model enzyme, since the adaptation of this enzyme towards new substrates is known to occur via the generation of this type of mutation. The results show that the mutagenesis method presented here is an effective tool for accessing formerly unexplorable sequence space and can contribute to the success of future directed evolution experiments.
Figures


References
-
- Myers R.M., Lerman,L.S. and Maniatis,T. (1985) A general method for saturation mutagenesis of cloned DNA fragments. Science, 229, 242–247. - PubMed
-
- Cox E.C. (1976) Bacterial mutator genes and the control of spontaneous mutation. Annu. Rev. Genet., 10, 135–156. - PubMed
-
- Bornscheuer U.T., Altenbuchner,J. and Meyer,H.H. (1998) Directed evolution of an esterase for the stereoselective resolution of a key intermediate in the synthesis of epothilones. Biotechnol. Bioeng., 58, 554–559. - PubMed
-
- Leung D.W., Chen,E. and Goeddel,D.V. (1989) A method for random mutagenesis of a defined DNA segment using a modified polymerase chain reaction. Technique, 1, 11–15.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

