Reduced-median-network analysis of complete mitochondrial DNA coding-region sequences for the major African, Asian, and European haplogroups
- PMID: 11938495
- PMCID: PMC447592
- DOI: 10.1086/339933
Reduced-median-network analysis of complete mitochondrial DNA coding-region sequences for the major African, Asian, and European haplogroups
Erratum in
- Am J Hum Genet 2002 Aug;71(2):448-9
Abstract
The evolution of the human mitochondrial genome is characterized by the emergence of ethnically distinct lineages or haplogroups. Nine European, seven Asian (including Native American), and three African mitochondrial DNA (mtDNA) haplogroups have been identified previously on the basis of the presence or absence of a relatively small number of restriction-enzyme recognition sites or on the basis of nucleotide sequences of the D-loop region. We have used reduced-median-network approaches to analyze 560 complete European, Asian, and African mtDNA coding-region sequences from unrelated individuals to develop a more complete understanding of sequence diversity both within and between haplogroups. A total of 497 haplogroup-associated polymorphisms were identified, 323 (65%) of which were associated with one haplogroup and 174 (35%) of which were associated with two or more haplogroups. Approximately one-half of these polymorphisms are reported for the first time here. Our results confirm and substantially extend the phylogenetic relationships among mitochondrial genomes described elsewhere from the major human ethnic groups. Another important result is that there were numerous instances both of parallel mutations at the same site and of reversion (i.e., homoplasy). It is likely that homoplasy in the coding region will confound evolutionary analysis of small sequence sets. By a linkage-disequilibrium approach, additional evidence for the absence of human mtDNA recombination is presented here.
Figures
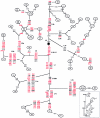
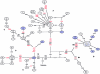
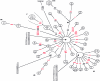
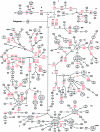
Comment in
-
Errors, phantoms and otherwise, in human mtDNA sequences.Am J Hum Genet. 2003 Jun;72(6):1585-6. doi: 10.1086/375406. Am J Hum Genet. 2003. PMID: 12817588 Free PMC article. No abstract available.
References
Electronic-Database Information
-
- Fluxus Engineering, http://fluxus-engineering.com/ (for Network 3.1 program)
-
- MitoKor, http://www.mitokor.com/science/560mtdnas.php (for the 560 mtDNA coding-region sequences)
-
- Sequence Analysis Server, http://genome.cs.mtu.edu/ (for the contig assembly program [CAP])
References
-
- Anderson S, Bankier AT, Barrell G, de Bruijn MHL, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, Schreier PH, Smith AJH, Staden R, Young IG (1981) Sequence and organization of the human mitochondrial genome. Nature 290:457–465 - PubMed
-
- Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N (1999) Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet 23:147 - PubMed
-
- Awadalla P, Eyre-Walker A, Maynard Smith J (1999) Linkage disequilibrium and recombination in hominid mitochondrial DNA. Science 286:2524–2525 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

