Dual roles of p300 in chromatin assembly and transcriptional activation in cooperation with nucleosome assembly protein 1 in vitro
- PMID: 11940655
- PMCID: PMC133748
- DOI: 10.1128/MCB.22.9.2974-2983.2002
Dual roles of p300 in chromatin assembly and transcriptional activation in cooperation with nucleosome assembly protein 1 in vitro
Abstract
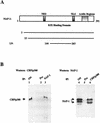
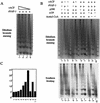
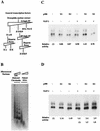
In a yeast two-hybrid screen to identify proteins that bind to the KIX domain of the coactivator p300, we obtained cDNAs encoding nucleosome assembly protein 1 (NAP-1), a 60-kDa histone H2A-H2B shuttling protein that promotes histone deposition. p300 associates preferentially with the H2A-H2B-bound form of NAP-1 rather than with the unbound form of NAP-1. Formation of NAP-1-p300 complexes was found to increase during S phase, suggesting a potential role for p300 in chromatin assembly. In micrococcal nuclease and supercoiling assays, addition of p300 promoted efficient chromatin assembly in vitro in conjunction with NAP-1 and ATP-utilizing chromatin assembly and remodeling factor; this effect was dependent in part on the intrinsic histone acetyltransferase activity of p300. Surprisingly, NAP-1 potently inhibited acetylation of core histones by p300, suggesting that efficient assembly requires acetylation of either NAP-1 or p300 itself. As p300 acted cooperatively with NAP-1 in stimulating transcription from a chromatin template in vitro, our results suggest a dual role of NAP-1-p300 complexes in promoting chromatin assembly and transcriptional activation.
Figures


References
-
- Adams, C. R., and R. T. Kamakaka. 1999. Chromatin assembly: biochemical identities and genetic redundancy. Curr. Opin. Genet. Dev. 9:185-190. - PubMed
-
- Akimaru, H., Y. Chen, P. Dai, D. X. Hou, M. Nonaka, S. M. Smolik, S. Armstrong, R. H. Goodman, and S. Ishii. 1997. Drosophila CBP is a coactivator of cubitus interruptus in hedgehog signalling. Nature 386:735-738. - PubMed
-
- Arias, J., A. S. Alberts, P. Brindle, F. X. Claret, T. Smeal, M. Karin, J. Feramisco, and M. Montminy. 1994. Activation of cAMP and mitogen responsive genes relies on a common nuclear factor. Nature 370:226-228. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
