LIS1, CLIP-170's key to the dynein/dynactin pathway
- PMID: 11940666
- PMCID: PMC133759
- DOI: 10.1128/MCB.22.9.3089-3102.2002
LIS1, CLIP-170's key to the dynein/dynactin pathway
Abstract
CLIP-170 is a plus-end tracking protein which may act as an anticatastrophe factor. It has been proposed to mediate the association of dynein/dynactin to microtubule (MT) plus ends, and it also binds to kinetochores in a dynein/dynactin-dependent fashion, both via its C-terminal domain. This domain contains two zinc finger motifs (proximal and distal), which are hypothesized to mediate protein-protein interactions. LIS1, a protein implicated in brain development, acts in several processes mediated by the dynein/dynactin pathway by interacting with dynein and other proteins. Here we demonstrate colocalization and direct interaction between CLIP-170 and LIS1. In mammalian cells, LIS1 recruitment to kinetochores is dynein/dynactin dependent, and recruitment there of CLIP-170 is dependent on its site of binding to LIS1, located in the distal zinc finger motif. Overexpression of CLIP-170 results in a zinc finger-dependent localization of a phospho-LIS1 isoform and dynactin to MT bundles, raising the possibility that CLIP-170 and LIS1 regulate dynein/dynactin binding to MTs. This work suggests that LIS1 is a regulated adapter between CLIP-170 and cytoplasmic dynein at sites involved in cargo-MT loading, and/or in the control of MT dynamics.
Figures



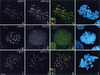
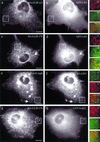
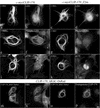

References
-
- Akhmanova, A., C. C. Hoogenraad, K. Drabek, T. Stepanova, B. Dortland, T. Verkerk, W. Vermeulen, B. M. Burgering, C. I. De Zeeuw, F. Grosveld, and N. Galjart. 2001. Clasps are CLIP-115 and -170 associating proteins involved in the regional regulation of microtubule dynamics in motile fibroblasts. Cell 104:923-935. - PubMed
-
- Allan, V. 1996. Motor proteins: a dynamic duo. Curr. Biol. 6:630-633. - PubMed
-
- Banks, J. D., and R. Heald. 2001. Chromosome movement: dynein-out at the kinetochore. Curr. Biol. 11:R128-R31. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
