Detection of fast light-activated H+ release and M intermediate formation from proteorhodopsin
- PMID: 11943070
- PMCID: PMC103662
- DOI: 10.1186/1472-6793-2-5
Detection of fast light-activated H+ release and M intermediate formation from proteorhodopsin
Abstract
Background: Proteorhodopsin (pR) is a light-activated proton pump homologous to bacteriorhodopsin and recently discovered in oceanic gamma-proteobacteria. One perplexing difference between these two proteins is the absence in pR of homologues of bR residues Glu-194 and Glu-204. These two residues, along with Arg-82, have been implicated in light-activated fast H+ release to the extracellular medium in bR. It is therefore uncertain that pR carries out its physiological activity using a mechanism that is completely homologous to that of bR.
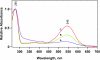
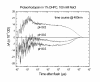
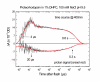
Results: A pR purification procedure is described that utilizes Phenylsepharose and hydroxylapatite columns and yields 85% (w/w) purity. Through SDS-PAGE of the pure protein, the molecular weight of E.-coli-produced pR was determined to be 36,000, approximately 9,000 more than the 27,000 predicted by the DNA sequence. Post-translational modification of one or more of the cysteine residues accounts for 5 kDa of the weight difference as measured on a cys-less pR mutant. At pH 9.5 and in the presence of octylglucoside and diheptanoylphosphotidylcholine, flash photolysis results in fast H+ release and a 400-nm absorbing (M-like) photoproduct. Both of these occur with a similar rise time (4-10 micros) as reported for monomeric bR in detergent.
Conclusions: The presence of fast H+ release in pR indicates that either different groups are responsible for fast H+ release in pR and bR (i.e. that the H+ release group is not highly conserved); or, that the H+ release group is conserved and is therefore likely Arg-94 itself in pR (and Arg-82 in bR, correspondingly).
Figures


References
-
- Béjà O, Aravind L, Koonin EV, Suzuki MT, Hadd A, Nguyen LP, Jovanovich SB, Gates CM, Feldman RA, Spudich JL, Spudich EN, DeLong EF. Bacterial rhodopsin: Evidence for a new type of phototrophy in the sea. Science. 2000;289:1902–1906. - PubMed
-
- Kouyama T, Nasuda-Kouyama A, Ikegami A, Mathew MK, Stoeckenius W. Bacteriorhodopsin photoreaction: identification of a long-lived intermediate N (P,R350) at high pH and its M-like photoproduct. Biochemistry. 1988;27:5855–5863. - PubMed
-
- Balashov SP, Imasheva E, Ebrey TG, Chen N, Menick DR, Crouch RK. Glutamate-194 to cysteine mutation inhibits fast light-induced-proton release in bacteriorhodopsin. Biochemistry. 1997;36:8671–8676. - PubMed
-
- Brown LS, Sasaki J, Kandori H, Maeda A, Needleman R, Lanyi JK. Glutamic acid 204 is the terminal proton release group at the extracellular surface of bacteriorhodopsin. J Biol Chem. 1995;270:27122–27126. - PubMed
-
- London E, Khorana HG. Denaturation and renaturation of bacteriorhodopsin in detergents and lipid-detergent mixtures. J Biol Chem. 1982;257:7003–7011. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

