Mechanism of plasmid-mediated quinolone resistance
- PMID: 11943863
- PMCID: PMC122823
- DOI: 10.1073/pnas.082092899
Mechanism of plasmid-mediated quinolone resistance
Abstract
Quinolones are potent antibacterial agents that specifically target bacterial DNA gyrase and topoisomerase IV. Widespread use of these agents has contributed to the rise of bacterial quinolone resistance. Previous studies have shown that quinolone resistance arises by mutations in chromosomal genes. Recently, a multiresistance plasmid was discovered that encodes transferable resistance to quinolones. We have cloned the plasmid-quinolone resistance gene, termed qnr, and found it in an integron-like environment upstream from qacE Delta 1 and sulI. The gene product Qnr was a 218-aa protein belonging to the pentapeptide repeat family and shared sequence homology with the immunity protein McbG, which is thought to protect DNA gyrase from the action of microcin B17. Qnr had pentapeptide repeat domains of 11 and 28 tandem copies, separated by a single glycine with a consensus sequence of A/C D/N L/F X X. Because the primary target of quinolones is DNA gyrase in Gram-negative strains, we tested the ability of Qnr to reverse the inhibition of gyrase activity by quinolones. Purified Qnr-His(6) protected Escherichia coli DNA gyrase from inhibition by ciprofloxacin. Gyrase protection was proportional to the concentration of Qnr-His(6) and inversely proportional to the concentration of ciprofloxacin. The protective activity of Qnr-His(6) was lost by boiling the protein and involved neither quinolone inactivation nor independent gyrase activity. Protection of topoisomerase IV, a secondary target of quinolone action in E. coli, was not evident. How Qnr protects DNA gyrase and the prevalence of this resistance mechanism in clinical isolates remains to be determined.
Figures
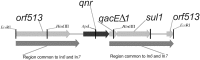







References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

