Evaluation of amplified rDNA restriction analysis (ARDRA) for the identification of cultured mycobacteria in a diagnostic laboratory
- PMID: 11945178
- PMCID: PMC101405
- DOI: 10.1186/1471-2180-2-4
Evaluation of amplified rDNA restriction analysis (ARDRA) for the identification of cultured mycobacteria in a diagnostic laboratory
Abstract
Background: The development of DNA amplification for the direct detection of M. tuberculosis from clinical samples has been a major goal of clinical microbiology during the last ten years. However, the limited sensitivity of most DNA amplification techniques restricts their use to smear positive samples. On the other hand, the development of automated liquid culture has increased the speed and sensitivity of cultivation of mycobacteria. We have opted to combine automated culture with rapid genotypic identification (ARDRA: amplified rDNA restriction analysis) for the detection resp. identification of all mycobacterial species at once, instead of attempting direct PCR based detection from clinical samples of M. tuberculosis only.
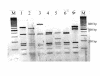
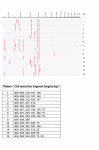
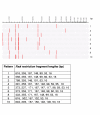
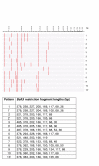
Results: During 1998-2000 a total of approx. 3500 clinical samples was screened for the presence of M. tuberculosis. Of the 151 culture positive samples, 61 were M. tuberculosis culture positive. Of the 30 smear positive samples, 26 were M. tuberculosis positive. All but three of these 151 mycobacterial isolates could be identified with ARDRA within on average 36 hours. The three isolates that could not be identified belonged to rare species not yet included in our ARDRA fingerprint library or were isolates with an aberrant pattern.
Conclusions: In our hands, automated culture in combination with ARDRA provides with accurate, practically applicable, wide range identification of mycobacterial species. The existing identification library covers most species, and can be easily updated when new species are studied or described. The drawback is that ARDRA is culture-dependent, since automated culture of M. tuberculosis takes on average 16.7 days (range 6 to 29 days). However, culture is needed after all to assess the antibiotic susceptibility of the strains.
Figures




References
-
- Wolinsky E. Mycobacterial diseases other than tuberculosis. Clin Infect Dis. 1992;15:1–12. - PubMed
-
- Espinal MA, Laserson K, Camacho M, Fusheng Z, Kim SJ, Tlali RE, Smith I, Suarez P, Antunes ML, George AG, et al. Determinants of drug-resistant tuberculosis: analysis of 11 countries. Int J Tubere Lung Dis. 2001;5:887–93. - PubMed
-
- Horsburg C, Metchok B, McGowen J, Thompson S. Clinical implications of recovery of M. avium complex from stool or respiratory tract of HIV infected individuals. AIDS. 1992;6:512–514. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

