Mutations in Saccharomyces cerevisiae gene SIR2 can have differential effects on in vivo silencing phenotypes and in vitro histone deacetylation activity
- PMID: 11950950
- PMCID: PMC102280
- DOI: 10.1091/mbc.01-10-0482
Mutations in Saccharomyces cerevisiae gene SIR2 can have differential effects on in vivo silencing phenotypes and in vitro histone deacetylation activity
Abstract
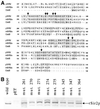
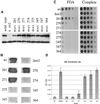
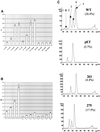
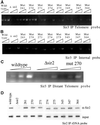
The yeast SIR2 gene and many of its homologs have been identified as NAD(+)-dependent histone deacetylases. To get a broader view of the relationship between the histone deacetylase activity of Sir2p and its in vivo functions we have mutated eight highly conserved residues in the core domain of SIR2. These mutations have a range of effects on the ability of Sir2p to deacetylate histones in vitro and to silence genes at the telomeres and HM loci. Interestingly, there is not a direct correlation between the in vitro and in vivo effects in some of these mutations. We also show that the histone deacetylase activity of Sir2p is necessary for the proper localiztion of the SIR complex to the telomeres.
Figures




References
-
- Aparicio OM, Billington BL, Gottschling DE. Modifiers of position effect are shared between telomeric and silent mating-type loci in S. cerevisiae. Cell. 1991;66:1279–1287. - PubMed
-
- Braunstein M, Rose AB, Holmes SG, Allis CD, Broach JR. Transcriptional silencing in yeast is associated with reduced nucleosome acetylation. Genes Dev. 1993;7:592–604. - PubMed
-
- Bryk M, Banerjee M, Murphy M, Knudsen KE, Garfinkel DJ, Curcio MJ. Transcriptional silencing of Ty1 elements in the RDN1 locus of yeast. Genes Dev. 1997;11:255–269. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

