Specific interaction of IP6 with human Ku70/80, the DNA-binding subunit of DNA-PK
- PMID: 11953323
- PMCID: PMC125973
- DOI: 10.1093/emboj/21.8.2038
Specific interaction of IP6 with human Ku70/80, the DNA-binding subunit of DNA-PK
Abstract
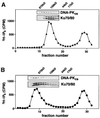
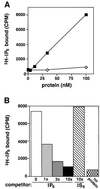
In eukaryotic cells, DNA double-strand breaks can be repaired by non-homologous end-joining, a process dependent upon Ku70/80, XRCC4 and DNA ligase IV. In mammals, this process also requires DNA-PK(cs), the catalytic subunit of the DNA-dependent protein kinase DNA-PK. Previously, inositol hexakisphosphate (IP6) was shown to be bound by DNA-PK and to stimulate DNA-PK-dependent end-joining in vitro. Here, we localize IP6 binding to the Ku70/80 subunits of DNA- PK, and show that DNA-PK(cs) alone exhibits no detectable affinity for IP6. Moreover, proteolysis mapping of Ku70/80 in the presence and absence of IP6 indicates that binding alters the conformation of the Ku70/80 heterodimer. The yeast homologue of Ku70/80, yKu70/80, fails to bind IP6, indicating that the function of IP6 in non-homologous end-joining, like that of DNA-PK(cs), is unique to the mammalian end-joining process.
Figures





References
-
- Chan D.W. and Lees-Miller,S.P. (1996) The DNA-dependent protein kinase is inactivated by autophosphorylation of the catalytic subunit. J. Biol. Chem., 271, 8936–8941. - PubMed
-
- Chan D.W., Ye,R.Q., Veillette,C.J. and Lees-Miller,S.P. (1999) DNA-dependent protein kinase phosphorylation sites in Ku70/80 heterodimer. Biochemistry, 38, 1819–1828. - PubMed
-
- Chu G. (1997) Double-strand break repair. J. Biol. Chem., 272, 24097–24100. - PubMed
-
- Critchlow S.E. and Jackson,S.P. (1998) DNA end-joining: from yeast to man. Trends Biochem. Sci., 23, 394–398. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

