Widespread distribution of a tet W determinant among tetracycline-resistant isolates of the animal pathogen Arcanobacterium pyogenes
- PMID: 11959557
- PMCID: PMC127165
- DOI: 10.1128/AAC.46.5.1281-1287.2002
Widespread distribution of a tet W determinant among tetracycline-resistant isolates of the animal pathogen Arcanobacterium pyogenes
Abstract
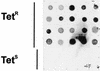
Tetracycline resistance is common among isolates of the animal commensal and opportunistic pathogen Arcanobacterium pyogenes. The tetracycline resistance determinant cloned from two bovine isolates of A. pyogenes was highly similar at the DNA level (92% identity) to the tet(W) gene, encoding a ribosomal protection tetracycline resistance protein, from the rumen bacterium Butyrivibrio fibrisolvens. The tet(W) gene was found in all 20 tetracycline-resistant isolates tested, indicating that it is a widely distributed determinant of tetracycline resistance in this organism. In 25% of tetracycline-resistant isolates, the tet(W) gene was associated with a mob gene, encoding a functional mobilization protein, and an origin of transfer, suggesting that the determinant may be transferable to other bacteria. In fact, low-frequency transfer of tet(W) was detected from mob+ A. pyogenes isolates to a tetracycline-sensitive A. pyogenes recipient. The mobile nature of this determinant and the presence of A. pyogenes in the gastrointestinal tract of cattle and pigs suggest that A. pyogenes may have inherited this determinant within the gastrointestinal tracts of these animals.
Figures



References
-
- Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. (ed.) 1994. Current protocols in molecular biology, vol. 1. Greene Publishing Associates and John Wiley and Sons, Inc., New York, N.Y.
-
- Bannam, T. L., P. K. Crellin, and J. I. Rood. 1995. Molecular genetics of the chloramphenicol-resistance transposon Tn4451 from Clostridium perfringens: the TnpX site specific recombinase excises a circular transposon molecule. Mol. Microbiol. 16:535-551. - PubMed
-
- Barbosa, T. M., K. P. Scott, and H. J. Flint. 1999. Evidence for recent intergeneric transfer of a new tetracycline resistance gene, tet(W), isolated from Butyrivibrio fibrisolvens, and the occurrence of tet(O) in ruminal bacteria. Environ. Microbiol. 1:53-64. - PubMed
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

