Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells
- PMID: 11959843
- PMCID: PMC152352
- DOI: 10.1101/gad.981002
Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells
Abstract
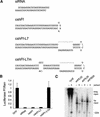
RNA interference (RNAi) was first recognized in Caenorhabditis elegans as a biological response to exogenous double-stranded RNA (dsRNA), which induces sequence-specific gene silencing. RNAi represents a conserved regulatory motif, which is present in a wide range of eukaryotic organisms. Recently, we and others have shown that endogenously encoded triggers of gene silencing act through elements of the RNAi machinery to regulate the expression of protein-coding genes. These small temporal RNAs (stRNAs) are transcribed as short hairpin precursors (approximately 70 nt), processed into active, 21-nt RNAs by Dicer, and recognize target mRNAs via base-pairing interactions. Here, we show that short hairpin RNAs (shRNAs) can be engineered to suppress the expression of desired genes in cultured Drosophila and mammalian cells. shRNAs can be synthesized exogenously or can be transcribed from RNA polymerase III promoters in vivo, thus permitting the construction of continuous cell lines or transgenic animals in which RNAi enforces stable and heritable gene silencing.
Figures

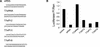
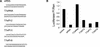
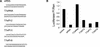



References
-
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001a;409:363–366. - PubMed
-
- Chong SS, Hu P, Hernandez N. Reconstitution of transcription from the human U6 small nuclear RNA promoter with eight recombinant polypeptides and a partially purified RNA polymerase III complex. J Biol Chem. 2001;276:20727–20734. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
