Stop codons affect 5' splice site selection by surveillance of splicing
- PMID: 11959978
- PMCID: PMC122760
- DOI: 10.1073/pnas.082095299
Stop codons affect 5' splice site selection by surveillance of splicing
Abstract
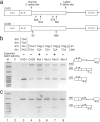
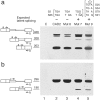
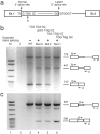
Pre-mRNA splicing involves recognition of a consensus sequence at the 5' splice site (SS). However, only some of the many potential sites that conform to the consensus are true ones, whereas the majority remain silent and are not normally used for splicing. We noticed that in most cases the utilization of such a latent intronic 5' SS for splicing would introduce an in-frame stop codon into the resultant mRNA. This finding suggested a link between SS selection and maintenance of an ORF within the mRNA. Here we tested this idea by analyzing the splicing of pre-mRNAs in which in-frame stop codons upstream of a latent 5' SS were mutated. We found that splicing with the latent site is indeed activated by such mutations. Our findings predict the existence of a checking mechanism, as a component of the nuclear pre-mRNA splicing machine, to ensure the maintenance of an ORF. This notion is highly important for accurate gene expression, as perturbations that would lead to splicing at these latent sites are expected to introduce in-frame stop codons into the majority of mRNAs.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

