Aggregation of proteins with expanded glutamine and alanine repeats of the glutamine-rich and asparagine-rich domains of Sup35 and of the amyloid beta-peptide of amyloid plaques
- PMID: 11960015
- PMCID: PMC122815
- DOI: 10.1073/pnas.042681599
Aggregation of proteins with expanded glutamine and alanine repeats of the glutamine-rich and asparagine-rich domains of Sup35 and of the amyloid beta-peptide of amyloid plaques
Abstract
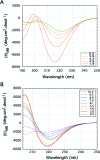
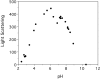
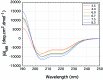
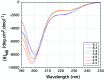
The exon-1 peptide of huntingtin has 51 Gln repeats and produces the symptoms of Huntington's disease in transgenic mice. Aggregation of the yeast Sup35 protein into prions has been attributed to its glutamine-rich and asparagine-rich domain. Here, we show that poly-L-asparagine forms polar zippers similar to those of poly-L-glutamine. In solution at acid pH, the glutamine-rich and asparagine-rich 18-residue Sup35 peptide, rendered soluble by the addition of two aspartates at the amino end and two lysines at the carboxyl end, gives a beta-sheet CD spectrum; it aggregates at neutral pH. A poly-alanine peptide D(2)A(10)K(2) gives an alpha-helical CD spectrum at all pHs and does not aggregate; a peptide with the sequence of the C-terminal helix of the alpha-chain of human hemoglobin, preceded by two aspartates and followed by two lysines, exhibits a random coil spectrum and does not aggregate either. Alignment of several beta-strands with the sequence of the 42-residue Alzheimer's amyloid beta-peptide shows that they can be linked together by a network of salt bridges. We also asked why single amino acid replacements can so destabilize the native structures of proteins that they unfold and form amyloids. The difference in free energy of a protein molecule between its native, fully ordered structure and an amorphous mixture of randomly coiled chains is only of the order of 10 kcal/mol. Theory shows that destabilization of the native structure by no more than 2 kcal/mol can increase the probability of nucleation of disordered aggregates from which amyloids could grow 130,000-fold.
Figures


References
-
- Williams R B. Science. 1994;264:566–569. - PubMed
-
- Patino M M, Liu J-J, Glover J R, Lindquist S. Science. 1996;273:622–626. - PubMed
-
- DePace A H, Santoso A, Milner O, Weissman J S. Cell. 1998;93:1241–1252. - PubMed
-
- Brais B, Bouchard J P, Xie Y G, Rochefort D L, Chretien N, Tome F M S, Lafreniere R G, Rommens J M, Uyama E, Nohira O, et al. Nat Genet. 1998;18:164–167. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

