A dynamin-like protein (ADL2b), rather than FtsZ, is involved in Arabidopsis mitochondrial division
- PMID: 11960028
- PMCID: PMC122839
- DOI: 10.1073/pnas.082663299
A dynamin-like protein (ADL2b), rather than FtsZ, is involved in Arabidopsis mitochondrial division
Abstract
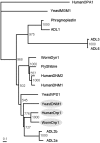
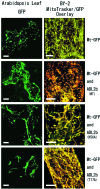
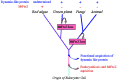
Recently, the FtsZ protein, which is known as a key component in bacterial cell division, was reported to be involved in mitochondrial division in algae. In yeast and animals, however, mitochondrial fission depends on the dynamin-like proteins Dnm1p and Drp1, respectively, whereas in green plants, no potential mitochondrial division genes have been identified. BLAST searches of the nuclear and mitochondrial genome sequences of Arabidopsis thaliana did not find any obvious homologue of the alpha-proteobacterial-type ftsZ genes. To determine whether mitochondrial division of higher plants depends on a dynamin-like protein, we cloned a cDNA for ADL2b, an Arabidopsis homologue of Dnm1p, and tested its subcellular localization and its dominant-negative effect on mitochondrial division. The fusion protein of green fluorescent protein and ADL2b was observed as punctate structures localized at the tips and at the constriction sites of mitochondria in live plant cells. Cells expressing dominant-negative mutant ADL2b proteins (K56A and T77F) showed a significant fusion, aggregation, and/or tubulation of mitochondria. We propose that mitochondrial division in higher plants is conducted by dynamin-like proteins similar to ADL2b in Arabidopsis. The evolutional points of loss of mitochondrial FtsZ and the functional acquisition of dynamin-like proteins in mitochondrial division are discussed.
Figures


References
-
- Gray M W. Int Rev Cytol. 1993;141:233–357. - PubMed
-
- Kuroiwa T, Kuroiwa H, Sakai A, Takahashi H, Toda K, Itoh R. Int Rev Cytol. 1998;181:1–41. - PubMed
-
- Bi E, Lutkenhaus J. Nature (London) 1991;354:161–164. - PubMed
-
- Osteryoung K W, McAndrew R S. Annu Rev Plant Phys Plant Mol Biol. 2001;52:315–333. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

