Regulation of mitochondrial D-loops by transcription factor A and single-stranded DNA-binding protein
- PMID: 11964388
- PMCID: PMC1084112
- DOI: 10.1093/embo-reports/kvf099
Regulation of mitochondrial D-loops by transcription factor A and single-stranded DNA-binding protein
Abstract
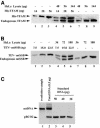
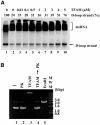
During replication, mitochondrial DNA (mtDNA) takes on a triple-stranded structure called a D-loop. Although their physiological roles are not understood, D-loops are implicated in replication and transcription of mtDNA. Little is known about the turnover of D-loops. We investigated the effects of mitochondrial transcription factor A (TFAM) and single-stranded DNA-binding protein (mtSSB) on D-loops. In human HeLa cells, TFAM and mtSSB are, respectively, 1700- and 3000-fold more abundant than mtDNA. This level of TFAM is two orders of magnitude higher than reported previously and is sufficient to wrap human mtDNA entirely. TFAM resolves D-loops in vitro if added in similar stoichiometries. mtSSB inhibits the resolution of mtDNA by TFAM but enhances resolution by RecG, a junction-specific helicase from Escherichia coli. Hence, mtSSB functions in both stabilization and resolution. We propose that TFAM and mtSSB are cooperatively involved in stabilizing D-loops and in the maintenance of mtDNA.
Figures



References
-
- Bogenhagen D. and Clayton, D.A. (1978) Mechanism of mitochondrial DNA replication in mouse L-cells: kinetics of synthesis and turnover of the initiation sequence. J. Mol. Biol., 119, 49–68. - PubMed
-
- Diffley J.F.X. and Stillman, B. (1992) DNA binding properties of an HMG1-related protein from yeast mitochondria. J. Biol. Chem., 267, 3368–3374. - PubMed
-
- Farr C.L., Wang, Y. and Kaguni, L.S. (1999) Functional interactions of mitochondrial DNA polymerase and single-stranded DNA-binding protein. J. Biol. Chem., 274, 14779–14785. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

