Mutational analysis of simian virus 40 T-antigen primosome activities in viral DNA replication
- PMID: 11967327
- PMCID: PMC136129
- DOI: 10.1128/jvi.76.10.5121-5130.2002
Mutational analysis of simian virus 40 T-antigen primosome activities in viral DNA replication
Abstract
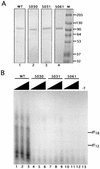
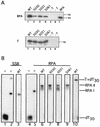
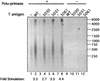
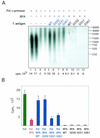
The recruitment of DNA polymerase alpha-primase (pol-prim) is a crucial step in the establishment of a functional replication complex in eukaryotic cells, but the mechanism of pol-prim loading and the composition of the eukaryotic primosome are poorly understood. In the model system for simian virus 40 (SV40) DNA replication in vitro, synthesis of RNA primers at the origin of replication requires only the viral tumor (T) antigen, replication protein A (RPA), pol-prim, and topoisomerase I. On RPA-coated single-stranded DNA (ssDNA), T antigen alone mediates priming by pol-prim, constituting a relatively simple primosome. T-antigen activities proposed to participate in its primosome function include DNA helicase and protein-protein interactions with RPA and pol-prim. To test the role of these activities of T antigen in mediating priming by pol-prim, three replication-defective T antigens with mutations in the ATPase or helicase domain have been characterized. All three mutant proteins interacted physically and functionally with RPA and pol-prim and bound ssDNA, and two of them displayed some helicase activity. However, only one of these, 5030, mediated primer synthesis and elongation by pol-prim on RPA-coated ssDNA. The results suggest that a novel activity, present in 5030 T antigen and absent in the other two mutants, is required for T-antigen primosome function.
Figures


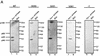
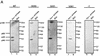
Similar articles
-
A specific docking site for DNA polymerase {alpha}-primase on the SV40 helicase is required for viral primosome activity, but helicase activity is dispensable.J Biol Chem. 2010 Oct 22;285(43):33475-33484. doi: 10.1074/jbc.M110.156240. Epub 2010 Aug 3. J Biol Chem. 2010. PMID: 20685648 Free PMC article.
-
Structure of a DNA polymerase alpha-primase domain that docks on the SV40 helicase and activates the viral primosome.J Biol Chem. 2010 May 28;285(22):17112-22. doi: 10.1074/jbc.M110.116830. Epub 2010 Mar 16. J Biol Chem. 2010. PMID: 20234039 Free PMC article.
-
The replication protein A binding site in simian virus 40 (SV40) T antigen and its role in the initial steps of SV40 DNA replication.J Virol. 1998 Dec;72(12):9771-81. doi: 10.1128/JVI.72.12.9771-9781.1998. J Virol. 1998. PMID: 9811712 Free PMC article.
-
Regulation of viral transcription and DNA replication by the SV40 large T antigen.Curr Top Microbiol Immunol. 1981;93:5-24. doi: 10.1007/978-3-642-68123-3_2. Curr Top Microbiol Immunol. 1981. PMID: 6269805 Review. No abstract available.
-
DNA replication: partners in the Okazaki two-step.Curr Biol. 2001 Oct 16;11(20):R842-4. doi: 10.1016/s0960-9822(01)00500-0. Curr Biol. 2001. PMID: 11676941 Review.
Cited by
-
Insights into hRPA32 C-terminal domain--mediated assembly of the simian virus 40 replisome.Nat Struct Mol Biol. 2005 Apr;12(4):332-9. doi: 10.1038/nsmb916. Epub 2005 Mar 27. Nat Struct Mol Biol. 2005. PMID: 15793585 Free PMC article.
-
Structural mechanism of RPA loading on DNA during activation of a simple pre-replication complex.EMBO J. 2006 Nov 29;25(23):5516-26. doi: 10.1038/sj.emboj.7601432. Epub 2006 Nov 16. EMBO J. 2006. PMID: 17110927 Free PMC article.
-
Methanosarcina acetivorans flap endonuclease 1 activity is inhibited by a cognate single-stranded-DNA-binding protein.J Bacteriol. 2006 Sep;188(17):6153-67. doi: 10.1128/JB.00045-06. J Bacteriol. 2006. PMID: 16923882 Free PMC article.
-
Role of the p68 subunit of human DNA polymerase alpha-primase in simian virus 40 DNA replication.Mol Cell Biol. 2002 Aug;22(16):5669-78. doi: 10.1128/MCB.22.16.5669-5678.2002. Mol Cell Biol. 2002. PMID: 12138179 Free PMC article.
-
Crystal structure of SV40 large T-antigen bound to p53: interplay between a viral oncoprotein and a cellular tumor suppressor.Genes Dev. 2006 Sep 1;20(17):2373-82. doi: 10.1101/gad.1456306. Genes Dev. 2006. PMID: 16951253 Free PMC article.
References
-
- Arezi, B., and R. D. Kuchta. 2000. Eukaryotic DNA primase. Trends Biochem. Sci. 25:572-576. - PubMed
-
- Beernink, H. T. H., and S. W. Morrical. 1999. RMPs: recombination/replication mediator proteins. Trends Biochem. Sci. 24:385-389. - PubMed
-
- Borowiec, J. A., F. B. Dean, P. A. Bullock, and J. Hurwitz. 1990. Binding and unwinding—how T antigen engages the SV40 origin of DNA replication. Cell 60:181-184. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

