New World arenavirus clade C, but not clade A and B viruses, utilizes alpha-dystroglycan as its major receptor
- PMID: 11967329
- PMCID: PMC136162
- DOI: 10.1128/jvi.76.10.5140-5146.2002
New World arenavirus clade C, but not clade A and B viruses, utilizes alpha-dystroglycan as its major receptor
Abstract
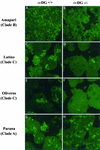
Alpha-dystroglycan (alpha-DG) has been identified as a major receptor for lymphocytic choriomeningitis virus (LCMV) and Lassa virus, two Old World arenaviruses. The situation with New World arenaviruses is less clear: previous studies demonstrated that Oliveros virus also exhibited high-affinity binding to alpha-DG but that Guanarito virus did not. To extend these initial studies, several additional Old and New World arenaviruses were screened for entry into mouse embryonic stem cells possessing or lacking alpha-DG. In addition, representative viruses were further analyzed for direct binding to alpha-DG by means of a virus overlay protein blot assay technique. These studies indicate that Old World arenaviruses use alpha-DG as a major receptor, whereas, of the New World arenaviruses, only clade C viruses (i.e., Oliveros and Latino viruses) use alpha-DG as a major receptor. New World clade A and B arenaviruses, which include the highly pathogenic Machupo, Guanarito, Junin, and Sabia viruses, appear to use a different receptor or coreceptor for binding. Previous studies with LCMV have suggested the need for a small aliphatic amino acid at LCMV GP1 glycoprotein amino acid position 260 to allow high-affinity binding to alpha-DG. As reported herein, this requirement appears to be broadly applicable to the arenaviruses as determined by more extensive analysis of alpha-DG receptor usage and GP1 sequences of Old and New World arenaviruses. In addition, GP1 amino acid position 259 also appears to be important, since all arenaviruses showing high-affinity alpha-DG binding possess a bulky aromatic amino acid (tyrosine or phenylalanine) at this position.
Figures



References
-
- Albariño, C. G., D. M. Posik, P. D. Ghiringhelli, M. E. Lozano, and V. Romanowski. 1998. Arenavirus phylogeny: a new insight. Virus Genes 16:39-46. - PubMed
-
- Baranowski, E., C. M. Ruiz-Jarabo, and E. Domingo. 2001. Evolution of cell recognition by viruses. Science 292:1102-1105. - PubMed
-
- Berger, E. A., P. M. Murphy, and J. M. Farber. 1999. Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism, and disease. Annu. Rev. Immunol. 17:657-700. - PubMed
-
- Bowen, M. D., C. J. Peters, and S. T. Nichol. 1996. The phylogeny of New World (Tacaribe complex) arenaviruses. Virology 219:285-290. - PubMed
-
- Bowen, M. D., C. J. Peters, and S. T. Nichol. 1997. Phylogenetic analysis of the Arenaviridae: patterns of virus evolution and evidence for co-speciations between arenaviruses and their rodent hosts. Mol. Phylogenet. Evol. 8:301-316. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

