Relationship between vaccinia virus intracellular cores, early mRNAs, and DNA replication sites
- PMID: 11967332
- PMCID: PMC136133
- DOI: 10.1128/jvi.76.10.5167-5183.2002
Relationship between vaccinia virus intracellular cores, early mRNAs, and DNA replication sites
Abstract
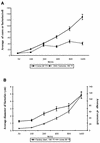
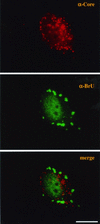
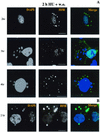
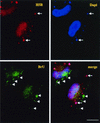
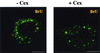
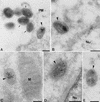
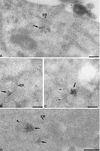
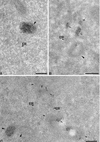
Virus assembly, a late event in the life cycle of vaccinia virus (VV), is preceded by a number of steps that all occur in the cytoplasm of the infected host cell: virion entry, delivery of the viral core into the cytoplasm, and transcription from these cores of early mRNAs, followed by the process of DNA replication. In the present study the quantitative and structural relationships between these distinct steps of VV morphogenesis were investigated. We show that viral RNA and DNA synthesis increases linearly with increasing amounts of incoming cores. Moreover, at multiplicities of infection that result in 10 to 40 cores per cell, an approximately 1:1 ratio between cores and sites of DNA replication exists, suggesting that each core is infectious. We have shown previously that VV early mRNAs collect in distinct granular structures that recruit components of the host cell translation machinery. Strikingly, these structures appeared to form some distance away from intracellular cores (M. Mallardo, S. Schleich, and J. Krijnse Locker, Mol. Biol. Cell 12:3875-3891, 2001). In the present study the intracellular locations of the sites of early mRNA accumulation and those of the subsequent process of DNA replication were compared. We show that these are distinct structures that have different intracellular locations. Finally, we study the fate of the parental DNA after core uncoating. By electron microscopy, cores were found close to membranes of the endoplasmic reticulum (ER) and the parental DNA, once it had left the core, appeared to associate preferentially with the cytosolic side of those membranes. Since we have previously shown that the process of DNA replication occurs in an ER-enclosed cytosolic "subcompartment" (N. Tolonen, L. Doglio, S. Schleich, and J. Krijnse Locker, Mol. Biol. Cell 12:2031-2046, 2001), the present data suggest that the parental DNA is released into the cytosol and associates with the same membranes where DNA replication is subsequently initiated. The combined data are discussed with respect to the cytosolic organization of VV morphogenesis.
Figures

Similar articles
-
The Vaccinia virus E8R gene product: a viral membrane protein that is made early in infection and packaged into the virions' core.J Virol. 2002 Oct;76(19):9773-86. doi: 10.1128/jvi.76.19.9773-9786.2002. J Virol. 2002. PMID: 12208956 Free PMC article.
-
Microtubule-dependent organization of vaccinia virus core-derived early mRNAs into distinct cytoplasmic structures.Mol Biol Cell. 2001 Dec;12(12):3875-91. doi: 10.1091/mbc.12.12.3875. Mol Biol Cell. 2001. PMID: 11739787 Free PMC article.
-
The vaccinia virus I3L gene product is localized to a complex endoplasmic reticulum-associated structure that contains the viral parental DNA.J Virol. 2003 May;77(10):6014-28. doi: 10.1128/jvi.77.10.6014-6028.2003. J Virol. 2003. PMID: 12719593 Free PMC article.
-
The mode of assembly of alphavirus cores implies a mechanism for the disassembly of the cores in the early stages of infection. Brief review.Arch Virol. 1987;94(1-2):1-14. doi: 10.1007/BF01313721. Arch Virol. 1987. PMID: 3034197 Review. No abstract available.
-
Virus synthesis and replication: reovirus vs. vaccinia virus.Yale J Biol Med. 1980 Jan-Feb;53(1):27-39. Yale J Biol Med. 1980. PMID: 6990634 Free PMC article. Review.
Cited by
-
Single-cell analysis of VACV infection reveals pathogen-driven timing of early and late phases and host-limited dynamics of virus production.PLoS Pathog. 2024 Aug 2;20(8):e1012423. doi: 10.1371/journal.ppat.1012423. eCollection 2024 Aug. PLoS Pathog. 2024. PMID: 39093901 Free PMC article.
-
DNA virus replication compartments.J Virol. 2014 Feb;88(3):1404-20. doi: 10.1128/JVI.02046-13. Epub 2013 Nov 20. J Virol. 2014. PMID: 24257611 Free PMC article. Review.
-
The block in assembly of modified vaccinia virus Ankara in HeLa cells reveals new insights into vaccinia virus morphogenesis.J Virol. 2002 Aug;76(16):8318-34. doi: 10.1128/jvi.76.16.8318-8334.2002. J Virol. 2002. PMID: 12134037 Free PMC article.
-
The Vaccinia virus E8R gene product: a viral membrane protein that is made early in infection and packaged into the virions' core.J Virol. 2002 Oct;76(19):9773-86. doi: 10.1128/jvi.76.19.9773-9786.2002. J Virol. 2002. PMID: 12208956 Free PMC article.
-
Vaccinia virus, a promising new therapeutic agent for pancreatic cancer.Immunotherapy. 2015;7(12):1249-58. doi: 10.2217/imt.15.90. Epub 2015 Nov 23. Immunotherapy. 2015. PMID: 26595180 Free PMC article. Review.
References
-
- Allan, V. 1996. Role of motor proteins in organizing the endoplasmic reticulum and Golgi apparatus. Semin. Cell Dev. Biol. 7:335-342.
-
- Beaud, G., and R. Beaud. 1997. Preferential virosomal location of underphosphorylated H5R protein synthesized in vaccinia virus-infected cells. J. Gen. Virol. 78:3297-3302. - PubMed
-
- Cairns, H. J. F. 1960. The initiation of vaccinia infection. Virology 11:603-623. - PubMed
-
- Cooper, J. A., and B. Moss. 1979. In vitro translation of immediate early, early, and late classes of RNA from vaccinia virus-infected cells. Virology 96:368-380. - PubMed
-
- Dahl, R., and J. R. Kates. 1970. Synthesis of vaccinia virus ‘early’ and ‘late’ messenger RNA in vitro with nucleoprotein structures isolated from infected cells. Virology 42:463-472. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

