Direct comparison of binding equilibrium, thermodynamic, and rate constants determined by surface- and solution-based biophysical methods
- PMID: 11967359
- PMCID: PMC2373566
- DOI: 10.1110/ps.4330102
Direct comparison of binding equilibrium, thermodynamic, and rate constants determined by surface- and solution-based biophysical methods
Abstract
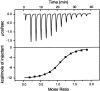
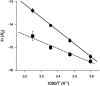
The binding interactions of small molecules with carbonic anhydrase II were used as model systems to compare the reaction constants determined from surface- and solution-based biophysical methods. Interaction data were collected for two arylsulfonamide compounds, 4-carboxybenzenesulfonamide (CBS) and 5-dimethyl-amino-1-naphthalene-sulfonamide (DNSA), binding to the enzyme using surface plasmon resonance, isothermal titration calorimetry, and stopped-flow fluorescence. We demonstrate that when the surface plasmon resonance biosensor experiments are performed with care, the equilibrium, thermodynamic, and kinetic constants determined from this surface-based technique match those acquired in solution. These results validate the use of biosensor technology to collect reliable data on small molecules binding to immobilized macromolecular targets. Binding kinetics were shown to provide more detailed information about complex formation than equilibrium constants alone. For example, although carbonic anhydrase II bound DNSA with twofold higher affinity than CBS, kinetic analysis revealed that CBS had a fourfold slower dissociation rate. Analysis of the binding and transition state thermodynamics also revealed significant differences in the enthalpy and entropy of complex formation. The lack of labeling requirements, high information content, and high throughput of surface plasmon resonance biosensors will make this technology an important tool for characterizing the interactions of small molecules with enzymes and receptors.
Figures






References
-
- Chegwidden, W.R. and Carter, N.D. 2000. Introduction to the carbonic anhydrases. In The carbonic anhydrases: New horizons (eds. W.R. Chegwidden, N.D. Carter, and Y.H. Edwards), pp. 13–28. Birkhauser Verlag, Basel.
-
- Hämäläinen, M.D., Markgren, P.-O., Schaal, W., Karlén, A., Classon, B., Vrang, L., Samuelsson, B., Hallberg, A., and Danielson, U.H. 2000. Characterization of a set of HIV-1 protease inhibitors using binding kinetics data from a biosensor-based screen. J. Biomol. Screen. 5 353–360. - PubMed
-
- Johnsson, B., Löfås, S., and Lindquist, G. 1991. Immobilization of proteins to a carboxymethyldextran-modified gold surface for biospecific interaction analysis in surface plasmon resonance sensors. Anal. Biochem. 198 268–277. - PubMed
-
- Kampranis, S.C., Gormley, N.A., Tranter, R., Orphanides, G., and Maxwell, A. 1999. Probing the binding of coumarins and cyclothialidines to DNA gyrase. Biochemisty 38 1967–1976. - PubMed
-
- Karlsson, R., Kullman-Magnusson, M., Hämäläinen, M.D., Remaeus, A., Andersson, K., Borg, P., Gyzander, E., and Deinum, J. 2000. Biosensor analysis of drug–target interactions: Direct and competitive binding assays for investigation of interactions between thrombin and thrombin inhibitors. Anal. Biochem. 278 1–13. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

