Genes for two mitochondrial ribosomal proteins in flowering plants are derived from their chloroplast or cytosolic counterparts
- PMID: 11971146
- PMCID: PMC150693
- DOI: 10.1105/tpc.010483
Genes for two mitochondrial ribosomal proteins in flowering plants are derived from their chloroplast or cytosolic counterparts
Abstract
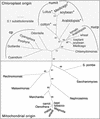
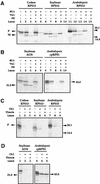
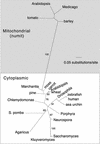
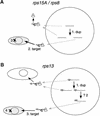
Often during flowering plant evolution, ribosomal protein genes have been lost from the mitochondrion and transferred to the nucleus. Here, we show that substitution by a duplicated, divergent gene originally encoding the chloroplast or cytosolic ribosomal protein counterpart accounts for two missing mitochondrial genes in diverse angiosperms. The rps13 gene is missing from the mitochondrial genome of many rosids, and a transferred copy of this gene is not evident in the nucleus of Arabidopsis, soybean, or cotton. Instead, these rosids contain a divergent nuclear copy of an rps13 gene of chloroplast origin. The product of this gene from all three rosids was shown to be imported into isolated mitochondria but not into chloroplasts. The rps8 gene is missing from the mitochondrion and nucleus of all angiosperms examined. A divergent copy of the gene encoding its cytosolic counterpart (rps15A) was identified in the nucleus of four angiosperms and one gymnosperm. The product of this gene from Arabidopsis and tomato was imported successfully into mitochondria. We infer that rps13 was lost from the mitochondrial genome and substituted by a duplicated nuclear gene of chloroplast origin early in rosid evolution, whereas rps8 loss and substitution by a gene of nuclear/cytosolic origin occurred much earlier, in a common ancestor of angiosperms and gymnosperms.
Figures




References
-
- Adams, K.L., Daley, D.O., Qiu, Y.-L., Whelan, J., and Palmer, J.D. (2000). Repeated, recent and diverse transfers of a mitochondrial gene to the nucleus in flowering plants. Nature 408, 354–357. - PubMed
-
- Adams, K.L., Ong, H.C., and Palmer, J.D. (2001. a). Mitochondrial gene transfer in pieces: Fission of a ribosomal protein gene and partial or complete gene transfer to the nucleus. Mol. Biol. Evol. 18, 2289–2297. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

