Hemagglutinin sequence clusters and the antigenic evolution of influenza A virus
- PMID: 11972025
- PMCID: PMC122937
- DOI: 10.1073/pnas.082110799
Hemagglutinin sequence clusters and the antigenic evolution of influenza A virus
Abstract
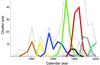
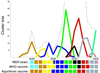
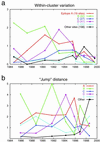
Continual mutations to the hemagglutinin (HA) gene of influenza A virus generate novel antigenic strains that cause annual epidemics. Using a database of 560 viral RNA sequences, we study the structure and tempo of HA evolution over the past two decades. We detect a critical length scale, in amino acid space, at which HA sequences aggregate into clusters, or swarms. We investigate the spatio-temporal distribution of viral swarms and compare it to the time series of the influenza vaccines recommended by the World Health Organization. We introduce a method for predicting future dominant HA amino acid sequences and discuss its potential relevance to vaccine choice. We also investigate the relationship between cluster structure and the primary antibody-combining regions of the HA protein.
Figures


References
-
- Hayden F G, Palese P. In: Clinical Virology. Richman D, Whitley R J, Hayden F G, editors. New York: Churchill Livingstone; 1997. pp. 911–942.
-
- Fitch W M, Bush R M, Bender C A, Subbarao K, Cox N J. J Hered. 2000;91:183–185. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

