Precision and functional specificity in mRNA decay
- PMID: 11972065
- PMCID: PMC122867
- DOI: 10.1073/pnas.092538799
Precision and functional specificity in mRNA decay
Abstract
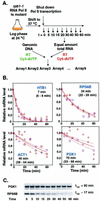
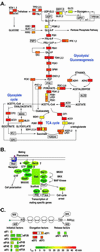
Posttranscriptional processing of mRNA is an integral component of the gene expression program. By using DNA microarrays, we precisely measured the decay of each yeast mRNA, after thermal inactivation of a temperature-sensitive RNA polymerase II. The half-lives varied widely, ranging from approximately 3 min to more than 90 min. We found no simple correlation between mRNA half-lives and ORF size, codon bias, ribosome density, or abundance. However, the decay rates of mRNAs encoding groups of proteins that act together in stoichiometric complexes were generally closely matched, and other evidence pointed to a more general relationship between physiological function and mRNA turnover rates. The results provide strong evidence that precise control of the decay of each mRNA is a fundamental feature of the gene expression program in yeast.
Figures


References
-
- Hargrove J L, Schmidt F H. FASEB J. 1989;3:2360–2370. - PubMed
-
- Cabrera C V, Lee J J, Ellison J W, Britten R J, Davidson E H. J Mol Biol. 1984;174:85–111. - PubMed
-
- Ross J. Trends Genet. 1996;12:171–175. - PubMed
-
- Casey J L, Hentze M W, Koeller D M, Caughman S W, Rouault T A, Klausner R D, Harford J B. Science. 1988;240:924–928. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

