Charge neutralization and DNA bending by the Escherichia coli catabolite activator protein
- PMID: 11972323
- PMCID: PMC113849
- DOI: 10.1093/nar/30.9.1879
Charge neutralization and DNA bending by the Escherichia coli catabolite activator protein
Abstract
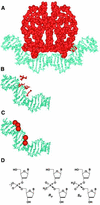
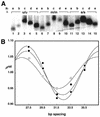
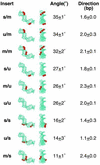
We are interested in the role of asymmetric phosphate neutralization in DNA bending induced by proteins. We describe an experimental estimate of the actual electrostatic contribution of asymmetric phosphate neutralization to the bending of DNA by the Escherichia coli catabolite activator protein (CAP), a prototypical DNA-bending protein. Following assignment of putative electrostatic interactions between CAP and DNA phosphates based on X-ray crystal structures, appropriate phosphates in the CAP half-site DNA were chemically neutralized by methylphosphonate substitution. DNA shape was then evaluated using a semi-synthetic DNA electrophoretic phasing assay. Our results confirm that the unmodified CAP DNA half-site sequence is intrinsically curved by 26 degrees in the direction enhanced in the complex with protein. In the absence of protein, neutralization of five appropriate phosphates increases DNA curvature to 32 degrees (approximately 23% increase), in the predicted direction. Shifting the placement of the neutralized phosphates changes the DNA shape, suggesting that sequence-directed DNA curvature can be modified by the asymmetry of phosphate neutralization. We suggest that asymmetric phosphate neutralization contributes favorably to DNA bending by CAP, but cannot account for the full DNA deformation.
Figures

References
-
- Liu-Johnson H.N., Gartenberg,M.R. and Crothers,D.M. (1986) The DNA binding domain and bending angle of the E. coli CAP protein. Cell, 47, 995–1005. - PubMed
-
- Schultz S.C., Shields,G.C. and Steitz,T.A. (1991) Crystal structure of a CAP-DNA complex: the DNA is bent by 90°. Science, 253, 1001–1007. - PubMed
-
- Parkinson G., Wilson,C., Gunasekera,A., Ebright,Y.W., Ebright,R.E. and Berman,H.M. (1996) Structure of the CAP-DNA complex at 2.5 Å resolution: a complete picture of the protein-DNA interface. J. Mol. Biol., 260, 395–408. - PubMed
-
- Kahn J.D. and Crothers,D.M. (1998) Measurement of the DNA bend angle induced by the catabolite activator protein using Monte Carlo simulation of cyclization kinetics. J. Mol. Biol., 276, 287–309. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

