Over-representation of repeats in stress response genes: a strategy to increase versatility under stressful conditions?
- PMID: 11972324
- PMCID: PMC113848
- DOI: 10.1093/nar/30.9.1886
Over-representation of repeats in stress response genes: a strategy to increase versatility under stressful conditions?
Abstract
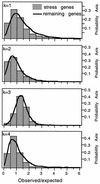
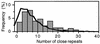
The survival of individual organisms facing stress is enhanced by the induction of a set of changes. As the intensity, duration and nature of stress is highly variable, the optimal response to stress may be unpredictable. To face such an uncertain future, it may be advantageous for a clonal population to increase its phenotypic heterogeneity (bet-hedging), ensuring that at least a subset of cells would survive the current stress. With current techniques, assessing the extent of this variability experimentally remains a challenge. Here, we use a bioinformatic approach to compare stress response genes with the rest of the genome for the presence of various kinds of repeated sequences, elements known to increase variability during the transfer of genetic information (i.e. during replication, but also during gene expression). We investigated the potential for illegitimate and homologous recombination of 296 Escherichia coli genes related to repair, recombination and physiological adaptations to different stresses. Although long repeats capable of engaging in homologous recombination are almost absent in stress response genes, we observed a significant high number of short close repeats capable of inducing phenotypic variability by slipped-mispair during DNA, RNA or protein synthesis.
Figures



References
-
- Kurland C.G. (1992) Translational accuracy and the fitness of bacteria. Annu. Rev. Genet., 26, 29–50. - PubMed
-
- Jorgensen F. and Kurland,C.G. (1990) Processivity errors of gene expression in Escherichia coli. J. Mol. Biol., 215, 511–521. - PubMed
-
- Rocha E.P.C., Danchin,A. and Viari,A. (1999) Functional and evolutionary roles of long repeats in prokaryotes. Res. Microbiol., 150, 725–733. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

